1C6Z
 
 | | ALTERNATE BINDING SITE FOR THE P1-P3 GROUP OF A CLASS OF POTENT HIV-1 PROTEASE INHIBITORS AS A RESULT OF CONCERTED STRUCTURAL CHANGE IN 80'S LOOP. | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PROTEIN (PROTEASE) | | Authors: | Munshi, S. | | Deposit date: | 1999-12-28 | | Release date: | 2000-12-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An alternate binding site for the P1-P3 group of a class of potent HIV-1 protease inhibitors as a result of concerted structural change in the 80s loop of the protease.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3D1X
 
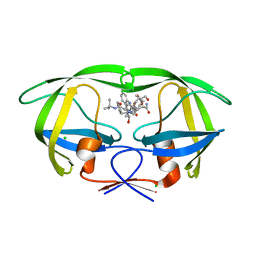 | | Crystal structure of HIV-1 mutant I54M and inhibitor saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-05-06 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
1C70
 
 | |
1C6Y
 
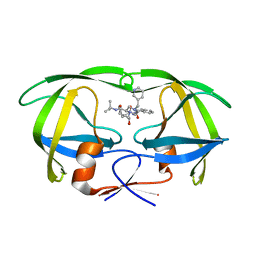 | |
3VZB
 
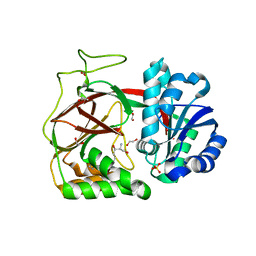 | | Crystal structure of Sphingosine Kinase 1 | | Descriptor: | (2S,3R,4E)-2-aminooctadec-4-ene-1,3-diol, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Min, X, Walker, N.P, Wang, Z. | | Deposit date: | 2012-10-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of sphingosine kinase 1 substrate recognition and catalysis.
Structure, 21, 2013
|
|
1OMX
 
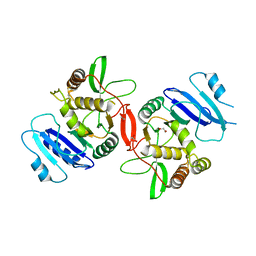 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2 | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1LBZ
 
 | | Crystal Structure of a complex (P32 crystal form) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus with 3 Calcium ions and Fructose-1,6 bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CALCIUM ION, fructose 1,6-bisphosphatase/inositol monophosphatase | | Authors: | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | Deposit date: | 2002-04-04 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
2Y1C
 
 | | X-ray structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with manganese. | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, MANGANESE (II) ION | | Authors: | Henriksson, L.M, Larsson, A.M.S, Bergfors, T, Bjorkelid, C, Unge, T, Mowbray, S.L, Jones, T.A. | | Deposit date: | 2010-12-08 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design, Synthesis and X-Ray Crystallographic Studies of Alpha-Aryl Substituted Fosmidomycin Analogues as Inhibitors of Mycobacterium Tuberculosis 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase
J.Med.Chem, 54, 2011
|
|
3VW7
 
 | | Crystal structure of human protease-activated receptor 1 (PAR1) bound with antagonist vorapaxar at 2.2 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Proteinase-activated receptor 1, ... | | Authors: | Zhang, C, Srinivasan, Y, Arlow, D.H, Fung, J.J, Palmer, D, Zheng, Y, Green, H.F, Pandey, A, Dror, R.O, Shaw, D.E, Weis, W.I, Coughlin, S.R, Kobilka, B.K. | | Deposit date: | 2012-08-07 | | Release date: | 2012-12-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution crystal structure of human protease-activated receptor 1
Nature, 492, 2012
|
|
3PZN
 
 | | Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol | | Descriptor: | CITRIC ACID, GLYCEROL, Mannan endo-1,4-beta-mannosidase. Glycosyl Hydrolase family 5 | | Authors: | Santos, C.R, Meza, A.N, Paiva, J.H, Silva, J.C, Ruller, R, Prade, R.A, Squina, F.M, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of a novel hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1
To be Published
|
|
3PO6
 
 | | Crystal structure of human carbonic anhydrase II with 6,7-Dimethoxy-1-methyl-3,4-dihydroisoquinoline-2(1H)-sulfonamide | | Descriptor: | (1R)-6,7-dimethoxy-1-methyl-3,4-dihydroisoquinoline-2(1H)-sulfonamide, (1S)-6,7-dimethoxy-1-methyl-3,4-dihydroisoquinoline-2(1H)-sulfonamide, ACETATE ION, ... | | Authors: | Mader, P, Brynda, J, Gitto, R, Agnello, S, Ferro, S, De Luca, L, Vullo, D, Supuran, C.T, Chimirri, A. | | Deposit date: | 2010-11-22 | | Release date: | 2011-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for the interaction between carbonic anhydrase and 1,2,3,4-tetrahydroisoquinolin-2-ylsulfonamides.
J.Med.Chem., 54, 2011
|
|
4FCE
 
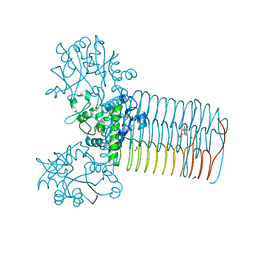 | | Crystal structure of Yersinia pestis GlmU in complex with alpha-D-glucosamine 1-phosphate (GP1) | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, Bifunctional protein GlmU, ... | | Authors: | Nocek, B, Kuhn, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Crystal structure of Yersinia pestis GlmU in complex with alpha-D-glucosamine 1-phosphate (GP1)
To be Published
|
|
4FL8
 
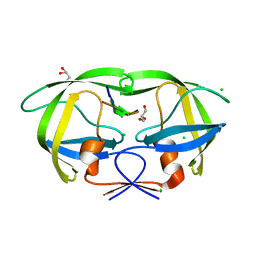 | | HIV-1 protease complexed with gem-diol-amine tetrahedral intermediate | | Descriptor: | CHLORIDE ION, GLYCEROL, HIV-1 protease, ... | | Authors: | Tie, Y.F, Shen, C.H, Weber, I.T. | | Deposit date: | 2012-06-14 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Capturing the Reaction Pathway in Near-Atomic-Resolution Crystal Structures of HIV-1 Protease.
Biochemistry, 51, 2012
|
|
4BHG
 
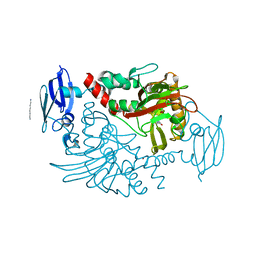 | | Three dimensional structure of human gamma-butyrobetaine hydroxylase in complex with 3-(1-Ethyl-1,1-dimethylhydrazin-1-ium-2-yl)propanoate | | Descriptor: | 3-(1-Ethyl-1,1-dimethylhydrazin-1-ium-2-yl)propanoic acid, GAMMA-BUTYROBETAINE DIOXYGENASE, HEXANE-1,6-DIAMINE, ... | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three Dimensional Structure of Human Gamma-Butyrobetaine Hydroxylase in Complex with 3-(1-Ethyl-1,1-Dimethylhydrazin-1-Ium-2-Yl)Propanoate
To be Published
|
|
2E40
 
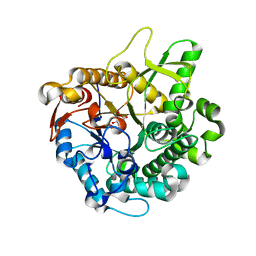 | | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium in complex with gluconolactone | | Descriptor: | Beta-glucosidase, D-glucono-1,5-lactone | | Authors: | Nijikken, Y, Tsukada, T, Igarashi, K, Samejima, M, Fushinobu, S. | | Deposit date: | 2006-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium
Febs Lett., 581, 2007
|
|
3PWM
 
 | | HIV-1 Protease Mutant L76V with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Zhang, Y, Weber, I.T. | | Deposit date: | 2010-12-08 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The L76V Drug Resistance Mutation Decreases the Dimer Stability and Rate of Autoprocessing of HIV-1 Protease by Reducing Internal Hydrophobic Contacts.
Biochemistry, 50, 2011
|
|
1UWA
 
 | | L290F mutant rubisco from chlamydomonas | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Taylor, T.C, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2004-02-03 | | Release date: | 2005-01-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Altered Intersubunit Interactions in Crystal Structures of Catalytically Compromised Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase
Biochemistry, 44, 2005
|
|
1GNO
 
 | | HIV-1 PROTEASE (WILD TYPE) COMPLEXED WITH U89360E (INHIBITOR) | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Treharne, A, Hartsuck, J.A, Foundling, S, Tang, J. | | Deposit date: | 1996-05-04 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of complexes of a peptidic inhibitor with wild-type and two mutant HIV-1 proteases.
Biochemistry, 35, 1996
|
|
5UEM
 
 | | Crystal structure of 354NC37 Fab in complex with HIV-1 clade AE strain 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 354NC37 Fab Heavy Chain, ... | | Authors: | Sievers, S.A, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-01-02 | | Release date: | 2018-01-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller.
Sci Transl Med, 9, 2017
|
|
3UDD
 
 | | Tankyrase-1 in complex with small molecule inhibitor | | Descriptor: | 3-(4-methoxyphenyl)-5-({[4-(4-methoxyphenyl)-5-methyl-4H-1,2,4-triazol-3-yl]sulfanyl}methyl)-1,2,4-oxadiazole, GLYCEROL, SULFATE ION, ... | | Authors: | Kirby, C.A, Stams, T. | | Deposit date: | 2011-10-28 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | [1,2,4]triazol-3-ylsulfanylmethyl)-3-phenyl-[1,2,4]oxadiazoles: antagonists of the Wnt pathway that inhibit tankyrases 1 and 2 via novel adenosine pocket binding.
J.Med.Chem., 55, 2012
|
|
3R02
 
 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 7-[(cis-4-aminocyclohexyl)amino]-5-bromo-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Xiang, Y, Hirth, B, Asmussen, G, Biemann, H.-P, Good, A, Fitzgerald, M, Gladysheva, T, Jancsics, K, Liu, J, Metz, M, Papoulis, A, Skerlj, R, Stepp, D.J, Wei, R.R. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3EE5
 
 | | Crystal structure of human M340H-Beta1,4-Galactosyltransferase-I (M340H-B4GAL-T1) in complex with GLCNAC-Beta1,3-Gal-Beta-Naphthalenemethanol | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2008-09-04 | | Release date: | 2009-01-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Deoxygenated Disaccharide Analogs as Specific Inhibitors of {beta}1-4-Galactosyltransferase 1 and Selectin-mediated Tumor Metastasis
J.Biol.Chem., 284, 2009
|
|
2ZBZ
 
 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R84A mutant) in complex with 1,25-dihydroxyvitamin D3 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Cytochrome P450-SU1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugimoto, H, Shinkyo, R, Hayashi, K, Yoneda, S, Yamada, M, Kamakura, M, Ikushiro, S, Shiro, Y, Sakaki, T. | | Deposit date: | 2007-10-30 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of CYP105A1 (P450SU-1) in Complex with 1alpha,25-Dihydroxyvitamin D3
Biochemistry, 47, 2008
|
|
1CQX
 
 | | Crystal structure of the flavohemoglobin from Alcaligenes eutrophus at 1.75 A resolution | | Descriptor: | 1-[GLYCEROLYLPHOSPHONYL]-2-[8-(2-HEXYL-CYCLOPROPYL)-OCTANAL-1-YL]-3-[HEXADECANAL-1-YL]-GLYCEROL, FLAVIN-ADENINE DINUCLEOTIDE, FLAVOHEMOPROTEIN, ... | | Authors: | Ermler, U, Siddiqui, R.A, Cramm, R, Friedrich, B. | | Deposit date: | 1999-08-12 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the flavohemoglobin from Alcaligenes eutrophus at 1.75 A resolution.
EMBO J., 14, 1995
|
|
1W2D
 
 | | Human Inositol (1,4,5)-trisphosphate 3-kinase complexed with Mn2+/ADP/Ins(1,3,4,5)P4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, INOSITOL-TRISPHOSPHATE 3-KINASE A, ... | | Authors: | Gonzalez, B, Schell, M.J, Irvine, R.F, Williams, R.L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure of a Human Inositol 1,4,5-Trisphosphate 3-Kinase; Substrate Binding Reveals Why It is not a Phosphoinositide 3-Kinase
Mol.Cell, 15, 2004
|
|
