4WR2
 
 | | Crystal structure of a putative pyrimidine-specific ribonucleoside hydrolase (RihA) Protein from Shewanella loihica PV-4 (SHEW_0697, Target PSI-029635) with divalent cation and PEG 400 bound at the active site | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, Pyrimidine-specific ribonucleoside hydrolase RihA | | Authors: | Himmel, D.M, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R.D, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a putative pyrimidine-specific ribonucleoside hydrolase (RihA) Protein from Shewanella loihica PV-4 (SHEW_0697, Target PSI-029635) with divalent cation and PEG 400 bound at the active site
To be published
|
|
1H41
 
 | | Pseudomonas cellulosa E292A alpha-D-glucuronidase mutant complexed with aldotriuronic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-alpha-D-glucopyranuronic acid, ALPHA-GLUCURONIDASE, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-09-25 | | Release date: | 2003-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Alpha-Glucuronidase,Glca67A,of Cellvibrio Japonicus Utilizes the Carboxylate and Methyl Groups of Aldobiouronic Acid as Important Substrate Recognition Determinants
J.Biol.Chem., 278, 2003
|
|
1RAK
 
 | | Bacterial cytosine deaminase D314S mutant bound to 5-fluoro-4-(S)-hydroxyl-3,4-dihydropyrimidine. | | Descriptor: | (4S)-5-FLUORO-4-HYDROXY-3,4-DIHYDROPYRIMIDIN-2(1H)-ONE, Cytosine deaminase, FE (III) ION, ... | | Authors: | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | Deposit date: | 2003-10-31 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
4X0V
 
 | | Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 | | Descriptor: | Beta-1,3-1,4-glucanase | | Authors: | Meng, D, Liu, X, Wang, X, Li, F, Feng, Y. | | Deposit date: | 2014-11-24 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Structural Insights into the Substrate Specificity of a Glycoside Hydrolase Family 5 Lichenase from Caldicellulosiruptor sp. F32
Biochem. J., 2017
|
|
4WRH
 
 | | AKR1C3 complexed with breakdown product of N-(tert-butyl)-2-(2-chloro-4-(((3-mercapto-5-methyl-4H-1,2,4-triazol-4-yl)amino)methyl)-6-methoxyphenoxy)acetamide | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(aminomethyl)-2-chloro-6-methoxyphenoxy]-N-tert-butylacetamide, 5-methyl-4H-1,2,4-triazole-3-thiol, ... | | Authors: | Squire, C.J, Flanagan, J.U, Yosaatmadja, Y. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Breakdown product of N-(tert-butyl)-2-(2-chloro-4-(((3-mercapto-5-methyl-4H-1,2,4-triazol-4-yl)amino)methyl)-6-methoxyphenoxy)acetamide trapped in active site of AKR1C3
To Be Published
|
|
1RCM
 
 | |
3PAH
 
 | | HUMAN PHENYLALANINE HYDROXYLASE CATALYTIC DOMAIN DIMER WITH BOUND ADRENALINE INHIBITOR | | Descriptor: | 4-[(1S)-1-hydroxy-2-(methylamino)ethyl]benzene-1,2-diol, FE (III) ION, PHENYLALANINE HYDROXYLASE | | Authors: | Erlandsen, H, Flatmark, T, Stevens, R.C. | | Deposit date: | 1998-08-20 | | Release date: | 1999-04-27 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analysis of the human phenylalanine hydroxylase catalytic domain with bound catechol inhibitors at 2.0 A resolution.
Biochemistry, 37, 1998
|
|
1H8V
 
 | | The X-ray Crystal Structure of the Trichoderma reesei Family 12 Endoglucanase 3, Cel12A, at 1.9 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Shaw, A, Ropp, T.H, Wu, S, Bott, R, Cameron, A.D, Stahlberg, J, Mitchinson, C, Jones, T.A. | | Deposit date: | 2001-02-16 | | Release date: | 2001-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-Ray Crystal Structure of the Trichoderma Reesei Family 12 Endoglucanase 3, Cel12A, at 1.9 A Resolution
J.Mol.Biol., 308, 2001
|
|
1H13
 
 | | Structure of a cold-adapted family 8 xylanase | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Van Petegem, F, Collins, T, Meuwis, M.A, Feller, G, Gerday, C, Van Beeumen, J. | | Deposit date: | 2002-07-02 | | Release date: | 2003-03-13 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Structure of a Cold-Adapted Family 8 Xylanase at 1.3 A Resolution: Structural Adaptations to Cold and Investigation of the Active Site
J.Biol.Chem., 278, 2003
|
|
3OVQ
 
 | | Crystal Structure of hRPE and D-Ribulose-5-Phospate Complex | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, FE (II) ION, RIBULOSE-5-PHOSPHATE, ... | | Authors: | Liang, W.G, Ouyang, S.Y, Shaw, N, Joachimiak, A, Zhang, R.G, Liu, Z.J. | | Deposit date: | 2010-09-17 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Conversion of D-ribulose 5-phosphate to D-xylulose 5-phosphate: new insights from structural and biochemical studies on human RPE
Faseb J., 25, 2011
|
|
1REA
 
 | |
3OWH
 
 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ZINC ION, ... | | Authors: | Pegan, S.D, Sturdy, M, Mesecar, A.D. | | Deposit date: | 2010-09-18 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
3OQP
 
 | |
4YYE
 
 | |
1H4H
 
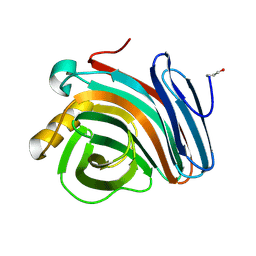 | | Oligosaccharide-binding to family 11 xylanases: both covalent intermediate and mutant-product complexes display 2,5B conformations at the active-centre | | Descriptor: | XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Sabini, E, Wilson, K.S, Danielsen, S, Schulein, M, Davies, G.J. | | Deposit date: | 2001-05-11 | | Release date: | 2002-05-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalysis and Specificity in Enzymatic Glycoside Hydrolysis: A 2,5B Conformation for the Glycosyl-Enzyme Intermediate Revealed by the Structure of the Bacillus Agaradhaerens Family 11 Xylanase.
Chem.Biol., 6, 1999
|
|
4ZE2
 
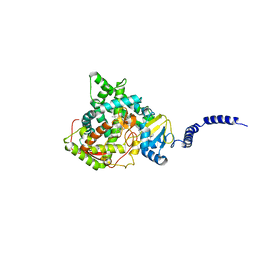 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) Y140H mutant complexed with itraconazole | | Descriptor: | 2-[(2R)-butan-2-yl]-4-{4-[4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazin-1-yl]phenyl}-2,4-dihydro-3H-1,2,4-triazol-3-one, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R, Monk, B.C, Tyndall, J.D.A. | | Deposit date: | 2015-04-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Triazole resistance mediated by mutations of a conserved active site tyrosine in fungal lanosterol 14 alpha-demethylase.
Sci Rep, 6, 2016
|
|
7FFR
 
 | |
1H5O
 
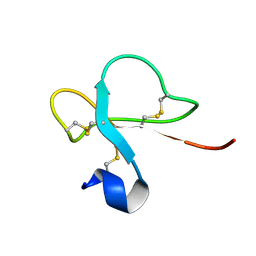 | | Solution structure of Crotamine, a neurotoxin from Crotalus durissus terrificus | | Descriptor: | MYOTOXIN | | Authors: | Nicastro, G, Franzoni, L, De Chiara, C, Mancin, C.A, Giglio, J.R, Spisni, A. | | Deposit date: | 2001-05-23 | | Release date: | 2003-05-09 | | Last modified: | 2013-07-24 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Crotamine, a Na+ Channel Affecting Toxin from Crotalus Durissus Terrificus Venom
Eur.J.Biochem., 270, 2003
|
|
4ZFC
 
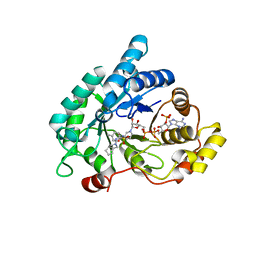 | | Crystal structure of AKR1C3 complexed with glicazide | | Descriptor: | Aldo-keto reductase family 1 member C3, N-[(3aR,6aS)-hexahydrocyclopenta[c]pyrrol-2(1H)-ylcarbamoyl]-4-methylbenzenesulfonamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zhrng, X, Hu, X. | | Deposit date: | 2015-04-21 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
1R34
 
 | | Golgi alpha-mannosidase II complex with 5-thio-D-mannopyranosylamidinium salt | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(1Z)-2-phenylethanimidoyl]-5-thio-alpha-D-mannopyranosylamine, ... | | Authors: | Kuntz, D.A, Xin, W, Kavelekar, L.M, Rose, D.R, Pinto, B.M. | | Deposit date: | 2003-09-30 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 5-Thio-d-glycopyranosylamines and their amidinium salts as potential transition-state mimics of glycosyl hydrolases: synthesis, enzyme inhibitory activities, X-ray crystallography, and molecular modeling
TETRAHEDRON ASYMMETRY, 16, 2005
|
|
1R43
 
 | | Crystal structure of beta-alanine synthase from Saccharomyces kluyveri (selenomethionine substituted protein) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-AMINO ISOBUTYRATE, ... | | Authors: | Lundgren, S, Gojkovic, Z, Piskur, J, Dobritzsch, D. | | Deposit date: | 2003-10-03 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Yeast beta-Alanine Synthase Shares a Structural Scaffold and Origin with Dizinc-dependent Exopeptidases
J.Biol.Chem., 278
|
|
7FFS
 
 | |
3OZ2
 
 | |
1RRM
 
 | |
4Z24
 
 | | Mimivirus R135 (residues 51-702) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GMC-type oxidoreductase R135 | | Authors: | Klose, T, Rossmann, M.G. | | Deposit date: | 2015-03-28 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mimivirus Enzyme that Participates in Viral Entry.
Structure, 23, 2015
|
|
