1I2X
 
 | | 2.4 A STRUCTURE OF A-DUPLEX WITH BULGED ADENOSINE, SPERMIDINE FORM | | Descriptor: | DNA/RNA (5'-R(*GP*CP*G)-D(P*AP*TP*AP*T)-R(P*AP*CP*GP*U)-3'), SPERMIDINE | | Authors: | Tereshko, V, Wallace, S, Usman, N, Wincott, F, Egli, M. | | Deposit date: | 2001-02-12 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic observation of "in-line" and "adjacent" conformations in a bulged self-cleaving RNA/DNA hybrid.
RNA, 7, 2001
|
|
1RPU
 
 | | Crystal Structure of CIRV p19 bound to siRNA | | Descriptor: | 19 kDa protein, 5'-R(P*CP*GP*UP*AP*CP*GP*CP*GP*UP*CP*AP*CP*GP*CP*GP*UP*AP*CP*GP*UP*U)-3' | | Authors: | Vargason, J.M, Szittya, G, Burgyan, J, Hall, T.M.T. | | Deposit date: | 2003-12-03 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Size selective recognition of siRNA by an RNA silencing suppressor
Cell(Cambridge,Mass.), 115, 2003
|
|
1M5L
 
 | |
1X9K
 
 | | An all-RNA Hairpin Ribozyme with mutation U39C | | Descriptor: | 5'-R(*AP*AP*UP*AP*GP*AP*GP*AP*AP*GP*CP*GP*A)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*GP*CP*AP*GP*UP*CP*CP*UP*AP*UP*U)-3', ... | | Authors: | Alam, S, Grum-Tokars, V, Krucinska, J, Kundracik, M.L, Wedekind, J.E. | | Deposit date: | 2004-08-21 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Conformational Heterogeneity at Position U37 of an All-RNA Hairpin Ribozyme with Implications for Metal Binding and the Catalytic Structure of the S-Turn.
Biochemistry, 44, 2005
|
|
1X9C
 
 | | An all-RNA Hairpin Ribozyme with mutation U39C | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | Alam, S, Grum-Tokars, V, Krucinska, J, Kundracik, M.L, Wedekind, J.E. | | Deposit date: | 2004-08-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Conformational Heterogeneity at Position U37 of an All-RNA Hairpin Ribozyme with Implications for Metal Binding and the Catalytic Structure of the S-Turn.
Biochemistry, 44, 2005
|
|
3C3L
 
 | |
1Q8N
 
 | | Solution Structure of the Malachite Green RNA Binding Aptamer | | Descriptor: | MALACHITE GREEN, RNA Aptamer | | Authors: | Flinders, J, DeFina, S.C, Brackett, D.M, Baugh, C, Wilson, C, Dieckmann, T. | | Deposit date: | 2003-08-21 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Recognition of planar and nonplanar ligands in the malachite green-RNA aptamer complex.
Chembiochem, 5, 2004
|
|
1JZC
 
 | |
4AOB
 
 | | SAM-I riboswitch containing the T. solenopsae Kt-23 in complex with S- adenosyl methionine | | Descriptor: | BARIUM ION, POTASSIUM ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Schroeder, K.T, Daldrop, P, McPhee, S.A, Lilley, D.M.J. | | Deposit date: | 2012-03-25 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure and Folding of a Rare, Natural Kink Turn in RNA with an Aa Pair at the 2B2N Position.
RNA, 18, 2012
|
|
1N8X
 
 | | Solution structure of HIV-1 Stem Loop SL1 | | Descriptor: | HIV-1 STEM LOOP SL1 MONOMERIC RNA | | Authors: | Lawrence, D.C, Stover, C.C, Noznitsky, J, Wu, Z, Summers, M.F. | | Deposit date: | 2002-11-21 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Intact Stem and Bulge of HIV-1 Psi-RNA Stem-Loop SL1
J.Mol.Biol., 326, 2003
|
|
4B5R
 
 | | SAM-I riboswitch bearing the H. marismortui K-t-7 | | Descriptor: | BARIUM ION, POTASSIUM ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Daldrop, P, Lilley, D.M.J. | | Deposit date: | 2012-08-07 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The Plasticity of a Structural Motif in RNA: Structural Polymorphism of a Kink Turn as a Function of its Environment.
RNA, 19, 2013
|
|
8ZDR
 
 | |
6M0T
 
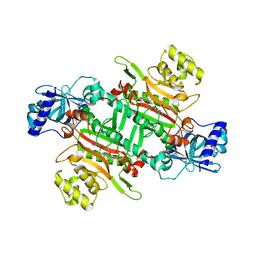 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Cladosporin derivative (CL-2) | | Descriptor: | (3R)-3-[(R)-[(2R,6S)-6-methyloxan-2-yl]-oxidanyl-methyl]-6,8-bis(oxidanyl)-3,4-dihydroisochromen-1-one, LYSINE, Lysine--tRNA ligase | | Authors: | Babbar, P, Sharma, A, Manickam, Y. | | Deposit date: | 2020-02-22 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Design, Synthesis, and Structural Analysis of Cladosporin-Based Inhibitors of Malaria Parasites.
Acs Infect Dis., 7, 2021
|
|
5AGH
 
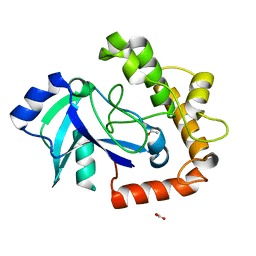 | | Crystal structure of the LeuRS editing domain of Candida albicans Mutant K510A | | Descriptor: | ACETATE ION, POTENTIAL CYTOSOLIC LEUCYL TRNA SYNTHETASE | | Authors: | Zhao, H, Palencia, A, Seiradake, E, Ghaemi, Z, Luthey-Schulten, Z, Cusack, S, Martinis, S.A. | | Deposit date: | 2015-02-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Analysis of the Resistance Mechanism of a Benzoxaborole Inhibitor Reveals Insight Into the Leucyl-tRNA Synthetase Editing Mechanism.
Acs Chem.Biol., 10, 2015
|
|
5AGI
 
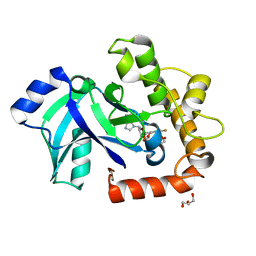 | | Crystal structure of the LeuRS editing domain of Candida albicans Mutant K510A in complex with the adduct formed by AN2690-AMP | | Descriptor: | GLYCEROL, POTENTIAL CYTOSOLIC LEUCYL TRNA SYNTHETASE, [(6-AMINO-9H-PURIN-9-YL)-[5-FLUORO-1,3-DIHYDRO-1-HYDROXY-2,1-BENZOXABOROLE]-4'YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Zhao, H, Palencia, A, Seiradake, E, Ghaemi, Z, Luthey-Schulten, Z, Cusack, S, Martinis, S.A. | | Deposit date: | 2015-02-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Analysis of the Resistance Mechanism of a Benzoxaborole Inhibitor Reveals Insight Into the Leucyl-tRNA Synthetase Editing Mechanism.
Acs Chem.Biol., 10, 2015
|
|
5AGJ
 
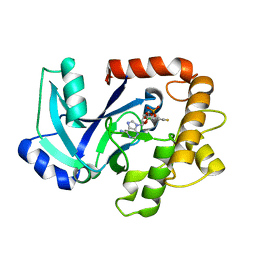 | | Crystal structure of the LeuRS editing domain of Candida albicans in complex with the adduct AN2690-AMP | | Descriptor: | POTENTIAL CYTOSOLIC LEUCYL TRNA SYNTHETASE, [(6-AMINO-9H-PURIN-9-YL)-[5-FLUORO-1,3-DIHYDRO-1-HYDROXY-2,1-BENZOXABOROLE]-4'YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Zhao, H, Palencia, A, Seiradake, E, Ghaemi, Z, Luthey-Schulten, Z, Cusack, S, Martinis, S.A. | | Deposit date: | 2015-02-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of the Resistance Mechanism of a Benzoxaborole Inhibitor Reveals Insight Into the Leucyl-tRNA Synthetase Editing Mechanism.
Acs Chem.Biol., 10, 2015
|
|
6AP0
 
 | |
6ANO
 
 | | Crystal structure of human FLASH N-terminal domain | | Descriptor: | CASP8-associated protein 2 | | Authors: | Aik, W.S, Tong, L. | | Deposit date: | 2017-08-14 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The N-terminal domains of FLASH and Lsm11 form a 2:1 heterotrimer for histone pre-mRNA 3'-end processing.
PLoS ONE, 12, 2017
|
|
6AOZ
 
 | |
1L3Z
 
 | | Crystal Structure Analysis of an RNA Heptamer | | Descriptor: | 5'-R(*GP*UP*AP*UP*AP*CP*A)-3', SODIUM ION | | Authors: | Shi, K, Pan, B, Sundaralingam, M. | | Deposit date: | 2002-03-04 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The crystal structure of an alternating RNA heptamer r(GUAUACA)
forming a six base-paired duplex with 3'-end adenine overhangs
Nucleic Acids Res., 31, 2003
|
|
2N0R
 
 | |
3H0M
 
 | | Structure of trna-dependent amidotransferase gatcab from aquifex aeolicus | | Descriptor: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, GLUTAMINE, Glutamyl-tRNA(Gln) amidotransferase subunit A, ... | | Authors: | Wu, J, Bu, W, Sheppard, K, Kitabatake, M, Soll, D, Smith, J.L. | | Deposit date: | 2009-04-09 | | Release date: | 2009-07-21 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into tRNA-Dependent Amidotransferase Evolution and Catalysis from the Structure of the Aquifex aeolicus Enzyme
J.Mol.Biol., 391, 2009
|
|
2P7F
 
 | | The Novel Use of a 2',5'-Phosphodiester Linkage as a Reaction Intermediate at the Active Site of a Small Ribozyme | | Descriptor: | COBALT HEXAMMINE(III), Loop A ribozyme strand, Loop B S-turn strand, ... | | Authors: | Torelli, A.T, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-03-20 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A comparison of vanadate to a 2'-5' linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization
Rna, 13, 2007
|
|
2P7E
 
 | | Vanadate at the Active Site of a Small Ribozyme Suggests a Role for Water in Transition-State Stabilization | | Descriptor: | 3' substrate strand, octameric fragment, 5' substrate strand, ... | | Authors: | Torelli, A.T, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-03-20 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A comparison of vanadate to a 2'-5' linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization
Rna, 13, 2007
|
|
3H0R
 
 | | Structure of trna-dependent amidotransferase gatcab from aquifex aeolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASPARAGINE, ... | | Authors: | Wu, J, Bu, W, Sheppard, K, Kitabatake, M, Soll, D, Smith, J.L. | | Deposit date: | 2009-04-10 | | Release date: | 2009-07-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights into tRNA-Dependent Amidotransferase Evolution and Catalysis from the Structure of the Aquifex aeolicus Enzyme
J.Mol.Biol., 391, 2009
|
|
