4H4K
 
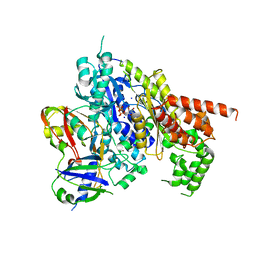 | | Structure of the Cmr2-Cmr3 subcomplex of the Cmr RNA-silencing complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cmr subunit Cmr2, CRISPR system Cmr subunit Cmr3, ... | | Authors: | Shao, Y, Cocozaki, A.I, Ramia, N.F, Terns, R.M, Terns, M.P, Li, H. | | Deposit date: | 2012-09-17 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structure of the cmr2-cmr3 subcomplex of the cmr RNA silencing complex.
Structure, 21, 2013
|
|
4U64
 
 | | Structure of the periplasmic output domain of the Legionella pneumophila LapD ortholog CdgS9 in the apo state | | Descriptor: | Two component histidine kinase, GGDEF domain protein/EAL domain protein | | Authors: | Chatterjee, D, Cooley, R.B, Boyd, C.D, Mehl, R.A, O'Toole, G.A, Sondermann, H.S. | | Deposit date: | 2014-07-27 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Mechanistic insight into the conserved allosteric regulation of periplasmic proteolysis by the signaling molecule cyclic-di-GMP.
Elife, 3, 2014
|
|
8SWH
 
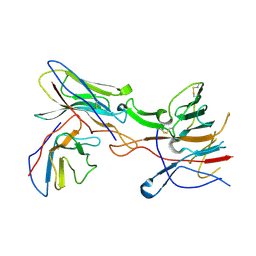 | |
4HAZ
 
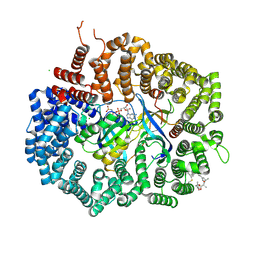 | | Crystal structure of CRM1 inhibitor Leptomycin B in complex with CRM1(R543S,K548E,K579Q)-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6NYB
 
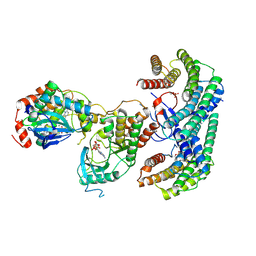 | | Structure of a MAPK pathway complex | | Descriptor: | 14-3-3 protein zeta, 5-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-6-carboxamide, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Park, E, Rawson, S, Li, K, Jeon, H, Eck, M.J. | | Deposit date: | 2019-02-11 | | Release date: | 2019-10-09 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes.
Nature, 575, 2019
|
|
8T42
 
 | | Model of TTLL6 MTBH1-2 bound to microtubule | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Mahalingan, K.K, Grotjahn, D, Li, Y, Lander, G.C, Zehr, E.A, Roll-Mecak, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for alpha-tubulin-specific and modification state-dependent glutamylation.
Nat.Chem.Biol., 2024
|
|
5A6F
 
 | | Cryo-EM structure of the Slo2.2 Na-activated K channel | | Descriptor: | GATING RING OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, RCK2 ELABORATION OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1 | | Authors: | Hite, R.K, Yuan, P, Li, Z, Hsuing, Y, Walz, T, MacKinnon, R. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-Electron Microscopy Structure of the Slo2.2 Na1-Activated K1 Channel
Nature, 527, 2015
|
|
5Z80
 
 | | Solution structure for the 1:1 complex of a platinum(II)-based tripod bound to a hybrid-1 human telomeric G-quadruplex | | Descriptor: | 4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]-N,N-bis[4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]phenyl]aniline, G-quadruplex DNA (26-MER) | | Authors: | Liu, W.T, Zhong, Y.F, Liu, L.Y, Zeng, W.J, Wang, F.Y, Yang, D.Z, Mao, Z.W. | | Deposit date: | 2018-01-30 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of multiple G-quadruplex complexes induced by a platinum(II)-based tripod reveal dynamic binding
Nat Commun, 9, 2018
|
|
4HAY
 
 | | Crystal structure of CRM1 inhibitor Leptomycin B in complex with CRM1(K548E,K579Q)-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exportin-1, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5ZAK
 
 | | Cryo-EM structure of human Dicer and its complexes with a pre-miRNA substrate | | Descriptor: | Endoribonuclease Dicer, RISC-loading complex subunit TARBP2 | | Authors: | Liu, Z, Wang, J, Cheng, H, Ke, X, Sun, L, Zhang, Q.C, Wang, H.-W. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-09 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate.
Cell, 173, 2018
|
|
5DT4
 
 | | Aurora A Kinase in Complex with AA35 and ATP in Space Group P6122 | | Descriptor: | 2-(3-bromophenyl)-8-fluoroquinoline-4-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, ... | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
5Z2O
 
 | |
3UO4
 
 | | Aurora A in complex with RPM1680 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-(biphenyl-2-ylamino)pyrimidin-2-yl]amino}benzoic acid, Aurora kinase A | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-11-16 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Novel Mechanism by Which Small Molecule Inhibitors Induce the DFG Flip in Aurora A.
Acs Chem.Biol., 7, 2012
|
|
4XHR
 
 | | Structure of a phospholipid trafficking complex, native | | Descriptor: | Mitochondrial distribution and morphology protein 35, Protein UPS1, mitochondrial | | Authors: | Yu, F, He, F, Wang, C, Zhang, P. | | Deposit date: | 2015-01-06 | | Release date: | 2015-07-01 | | Last modified: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of intramitochondrial phosphatidic acid transport mediated by Ups1-Mdm35 complex
Embo Rep., 16, 2015
|
|
3UP7
 
 | | Aurora A in complex with YL1-038-09 | | Descriptor: | 2-({2-[(4-carboxyphenyl)amino]pyrimidin-4-yl}amino)benzoic acid, Aurora kinase A | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-11-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Development of o-Chlorophenyl Substituted Pyrimidines as Exceptionally Potent Aurora Kinase Inhibitors.
J.Med.Chem., 55, 2012
|
|
5ZIA
 
 | |
7YQB
 
 | |
3UWD
 
 | | Crystal Structure of Phosphoglycerate Kinase from Bacillus Anthracis | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Zheng, H, Chruszcz, M, Porebski, P, Kudritska, M, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-01 | | Release date: | 2012-01-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structures of putative phosphoglycerate kinases from B. anthracis and C. jejuni.
J.Struct.Funct.Genom., 13, 2012
|
|
4HAT
 
 | |
5Z6L
 
 | |
4XEJ
 
 | | IRES bound to bacterial Ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhu, J, Korostelev, A, Noller, H.F, Donohue, J.P. | | Deposit date: | 2014-12-23 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Initiation of translation in bacteria by a structured eukaryotic IRES RNA.
Nature, 519, 2015
|
|
4XIZ
 
 | | Structure of a phospholipid trafficking complex with substrate | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, Mitochondrial distribution and morphology protein 35, Protein UPS1, ... | | Authors: | Yu, F, He, F, Wang, C, Zhang, P. | | Deposit date: | 2015-01-08 | | Release date: | 2015-07-01 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of intramitochondrial phosphatidic acid transport mediated by Ups1-Mdm35 complex
Embo Rep., 16, 2015
|
|
6NZ0
 
 | | Cryo-EM structure of AAV-2 in complex with AAVR PKD domains 1 and 2 | | Descriptor: | Capsid protein VP1, Dyslexia-associated protein KIAA0319-like protein, MAGNESIUM ION | | Authors: | Meyer, N.L, Xie, Q, Davulcu, O, Yoshioka, C, Chapman, M.S. | | Deposit date: | 2019-02-12 | | Release date: | 2019-06-12 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure of the gene therapy vector, adeno-associated virus with its cell receptor, AAVR.
Elife, 8, 2019
|
|
4UIS
 
 | | The cryoEM structure of human gamma-Secretase complex | | Descriptor: | GAMMA-SECRETASE, LYSOZYME | | Authors: | Sun, L, Zhao, L, Yang, G, Yan, C, Zhou, R, Zhou, X, Xie, T, Zhao, Y, Wu, S, Li, X, Shi, Y. | | Deposit date: | 2015-04-03 | | Release date: | 2015-06-10 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis of Human Gamma-Secretase Assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5DWP
 
 | | Crystal Structure of human transthyretin (TTR) processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | Transthyretin | | Authors: | Zander, U, Cornaciu, I, Forsyth, T, Yee, A, Marquez, J.A. | | Deposit date: | 2015-09-22 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Automated harvesting and processing of protein crystals through laser photoablation.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
