3SZX
 
 | | Crystal Structure of the Triplet Repeat in Myotonic Dystrophy Reveals Heterogeneous 1x1 Nucleotide UU Internal Loop Conformations | | Descriptor: | RNA (5'-R(P*UP*UP*GP*GP*GP*CP*CP*UP*GP*CP*UP*GP*CP*UP*GP*GP*UP*CP*C)-3') | | Authors: | Kumar, A, Park, H, Pengfei, F, Parkesh, R, Guo, M, Nettles, K.W, Disney, M.D. | | Deposit date: | 2011-07-19 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Crystal Structure of the Triplet Repeat in Myotonic Dystrophy Reveals Heterogeneous 1x1 Nucleotide UU Internal Loop Conformations
Biochemistry, 50, 2011
|
|
8TSV
 
 | |
8TQX
 
 | |
2LO6
 
 | | Structure of Nrd1 CID bound to phosphorylated RNAP II CTD | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, Protein NRD1 | | Authors: | Kubicek, K, Cerna, H, Pasulka, J, Holub, P, Hrossova, D, Loehr, F, Hofr, C, Vanacova, S, Stefl, R. | | Deposit date: | 2012-01-17 | | Release date: | 2012-12-26 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Serine phosphorylation and proline isomerization in RNAP II CTD control recruitment of Nrd1.
Genes Dev., 26, 2012
|
|
5JX4
 
 | | Crystal structure of E36-G37del mutant of the Bacillus caldolyticus cold shock protein. | | Descriptor: | Cold shock protein CspB, SULFATE ION | | Authors: | Carvajal, A, Castro-Fernandez, V, Cabrejos, D, Fuentealba, M, Pereira, H.M, Vallejos, G, Cabrera, R, Garratt, R.C, Komives, E.A, Ramirez-Sarmiento, C.A, Babul, J. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual dimerization of a BcCsp mutant leads to reduced conformational dynamics.
FEBS J., 284, 2017
|
|
1T5I
 
 | | Crystal structure of the C-terminal domain of UAP56 | | Descriptor: | C_TERMINAL DOMAIN OF A PROBABLE ATP-DEPENDENT RNA HELICASE | | Authors: | Zhao, R, Green, M.R, Shen, J. | | Deposit date: | 2004-05-04 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of UAP56, a "DEXD/H-box" protein involved in pre-mRNA splicing and mRNA export
Structure, 12, 2004
|
|
2D2K
 
 | | Crystal Structure of a minimal, native (U39) all-RNA hairpin ribozyme | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | Alam, S, Grum-Tokars, V, Krucinska, J, Kundracik, M.L, Wedekind, J.E. | | Deposit date: | 2005-09-11 | | Release date: | 2005-11-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Conformational heterogeneity at position U37 of an all-RNA hairpin ribozyme with implications for metal binding and the catalytic structure of the S-turn
Biochemistry, 44, 2005
|
|
2D2L
 
 | | Crystal Structure of a minimal, all-RNA hairpin ribozyme with a propyl linker (C3) at position U39 | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | Alam, S, Grum-Tokars, V, Krucinska, J, Kundracik, M.L, Wedekind, J.E. | | Deposit date: | 2005-09-11 | | Release date: | 2005-11-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational heterogeneity at position U37 of an all-RNA hairpin ribozyme with implications for metal binding and the catalytic structure of the S-turn
Biochemistry, 44, 2005
|
|
4Q5S
 
 | | Thermus thermophilus RNA polymerase initially transcribing complex containing 6-mer RNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*GP*CP*AP*GP*CP*CP*A)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3'), ... | | Authors: | Murakami, K.S. | | Deposit date: | 2014-04-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme.
J.Biol.Chem., 289, 2014
|
|
7QDD
 
 | |
8YGJ
 
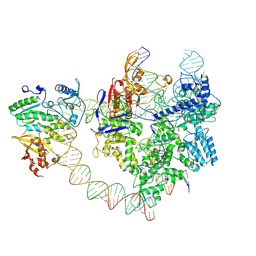 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 28-nt) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(P*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2024-02-26 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
4Q4Z
 
 | | Thermus thermophilus RNA polymerase de novo transcription initiation complex | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ADENOSINE-5'-TRIPHOSPHATE, DNA (25-MER), ... | | Authors: | Murakami, K.S. | | Deposit date: | 2014-04-15 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme.
J.Biol.Chem., 289, 2014
|
|
6L6V
 
 | | SPO1 Gp44 N-terminal region (1-55) | | Descriptor: | E3 protein | | Authors: | Liu, B, Wang, Z. | | Deposit date: | 2019-10-29 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Bacteriophage DNA Mimic Protein Employs a Non-specific Strategy to Inhibit the Bacterial RNA Polymerase.
Front Microbiol, 12, 2021
|
|
3AY0
 
 | | Crystal structure of Methanocaldococcus jannaschii Trm5 in complex with adenosine | | Descriptor: | ADENOSINE, Uncharacterized protein MJ0883, ZINC ION | | Authors: | Goto-Ito, S, Ito, T, Hou, Y.M, Yokoyama, S. | | Deposit date: | 2011-04-21 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Differentiating analogous tRNA methyltransferases by fragments of the methyl donor.
Rna, 17, 2011
|
|
6DTA
 
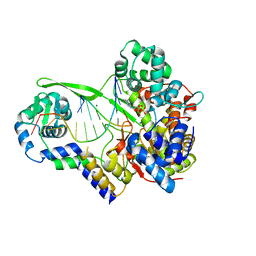 | | Bacteriophage N4 RNA polymerase II elongation complex 2 | | Descriptor: | DNA (5'-D(P*CP*CP*CP*AP*CP*CP*AP*AP*AP*AP*A)-3'), GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Molodtsov, V, Murakami, K.S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Minimalism and functionality: Structural lessons from the heterodimeric N4 bacteriophage RNA polymerase II.
J. Biol. Chem., 293, 2018
|
|
8YNP
 
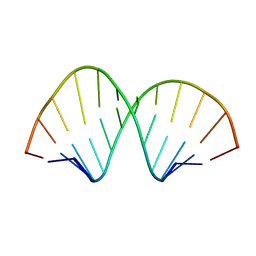 | | RNA duplex containing Methylformamide | | Descriptor: | RNA (5'-R(*GP*GP*AP*CP*(5BU)P*CP*GP*AP*GP*(HHX)P*CP*C)-3') | | Authors: | Kondo, J, Abe, H. | | Deposit date: | 2024-03-11 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Synthesis of 2'-formamidonucleoside phosphoramidites for suppressing the seed-based off-target effects of siRNAs.
Nucleic Acids Res., 52, 2024
|
|
7QDE
 
 | |
8YNO
 
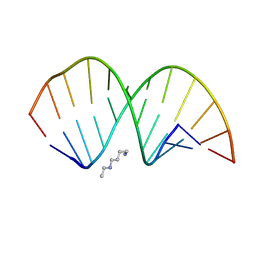 | | RNA duplex containing Formamide | | Descriptor: | RNA (5'-R(*GP*GP*AP*CP*(5BU)P*CP*GP*AP*GP*(HHU)P*CP*C)-3'), SPERMINE | | Authors: | Kondo, J, Abe, H. | | Deposit date: | 2024-03-11 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Synthesis of 2'-formamidonucleoside phosphoramidites for suppressing the seed-based off-target effects of siRNAs.
Nucleic Acids Res., 52, 2024
|
|
6DT8
 
 | | Bacteriophage N4 RNA polymerase II elongation complex 1 | | Descriptor: | DNA (5'-D(P*AP*CP*CP*CP*AP*CP*CP*AP*AP*AP*AP*A)-3'), RNA (5'-R(P*UP*GP*GP*UP*GP*G)-3'), RNAP1, ... | | Authors: | Molodtsov, V, Murakami, K.S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Minimalism and functionality: Structural lessons from the heterodimeric N4 bacteriophage RNA polymerase II.
J. Biol. Chem., 293, 2018
|
|
1D16
 
 | |
8VFS
 
 | |
6DT7
 
 | | Bacteriophage N4 RNA polymerase II and DNA complex | | Descriptor: | DNA (5'-D(P*CP*CP*AP*CP*CP*TP*GP*CP*A)-3'), RNAP1, RNAP2 | | Authors: | Molodtsov, V, Murakami, K.S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Minimalism and functionality: Structural lessons from the heterodimeric N4 bacteriophage RNA polymerase II.
J. Biol. Chem., 293, 2018
|
|
4HVC
 
 | | Crystal structure of human prolyl-tRNA synthetase in complex with halofuginone and ATP analogue | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, Bifunctional glutamate/proline--tRNA ligase, MAGNESIUM ION, ... | | Authors: | Zhou, H, Sun, L, Yang, X.L, Schimmel, P. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ATP-directed capture of bioactive herbal-based medicine on human tRNA synthetase.
Nature, 494, 2012
|
|
4PMI
 
 | | Crystal structure of Rev and Rev-response-element RNA complex | | Descriptor: | PHOSPHATE ION, Protein Rev, Rev-Response-Element RNA | | Authors: | Jayaraman, B, Crosby, D.C, Homer, C, Ribeiro, I, Mavor, D, Frankel, A.D. | | Deposit date: | 2014-05-21 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | RNA-directed remodeling of the HIV-1 protein Rev orchestrates assembly of the Rev-Rev response element complex.
Elife, 4, 2014
|
|
9HFL
 
 | |
