7UJT
 
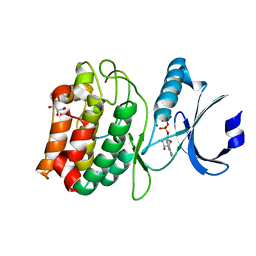 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and GluN2B(S1303D) in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Calcium/calmodulin-dependent protein kinase type II subunit alpha, GLYCEROL, ... | | Authors: | Ozden, C, Stratton, M.M, Garman, S.C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
7UJR
 
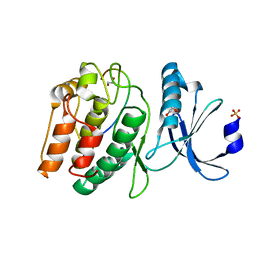 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and GluN2B | | Descriptor: | 1,2-ETHANEDIOL, Calcium/calmodulin-dependent protein kinase type II subunit alpha, Glutamate receptor ionotropic, ... | | Authors: | Ozden, C, Santos, N.J, Stratton, M.M, Garman, S.C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
7UJS
 
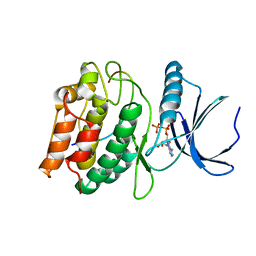 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and GluN2B in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Calcium/calmodulin-dependent protein kinase type II subunit alpha, Glutamate receptor ionotropic, ... | | Authors: | Ozden, C, Santos, N.J, Stratton, M.M, Garman, S.C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
7UJQ
 
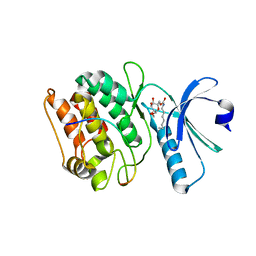 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and GluN2B | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha, GLYCEROL, Glutamate receptor ionotropic, ... | | Authors: | Ozden, C, Foster, J.C, Stratton, M.M, Garman, S.C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
7UJP
 
 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and GluN2B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Calcium/calmodulin-dependent protein kinase type II subunit alpha, ... | | Authors: | Ozden, C, Foster, J.C, Stratton, M.M, Garman, S.C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
1SNJ
 
 | | Solution structure of the DNA three-way junction with the A/C-stacked conformation | | Descriptor: | 36-MER | | Authors: | Wu, B, Girard, F, van Buuren, B, Schleucher, J, Tessari, M, Wijmenga, S. | | Deposit date: | 2004-03-11 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Global structure of a DNA three-way junction by solution NMR: towards prediction of 3H fold.
Nucleic Acids Res., 32, 2004
|
|
1SE9
 
 | |
1TN4
 
 | | FOUR CALCIUM TNC | | Descriptor: | CALCIUM ION, TROPONIN C | | Authors: | Love, M.L, Dominguez, R, Houdusse, A, Cohen, C. | | Deposit date: | 1997-09-18 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of four Ca2+-bound troponin C at 2.0 A resolution: further insights into the Ca2+-switch in the calmodulin superfamily.
Structure, 5, 1997
|
|
1U16
 
 | | Crystal structure of a duck-delta-crystallin-1 double loop mutant (DLM) in complex with sulfate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Delta crystallin I, ... | | Authors: | Tsai, M, Sampaleanu, L.M, Greene, C, Creagh, L, Haynes, C, Howell, P.L. | | Deposit date: | 2004-07-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A duck delta1 crystallin double loop mutant provides insight into residues important for argininosuccinate lyase activity.
Biochemistry, 43, 2004
|
|
1U15
 
 | | Crystal structure of a duck-delta-crystallin-1 double loop mutant (DLM) | | Descriptor: | Delta crystallin I | | Authors: | Tsai, M, Sampaleanu, L.M, Greene, C, Creagh, L, Haynes, C, Howell, P.L. | | Deposit date: | 2004-07-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A duck delta1 crystallin double loop mutant provides insight into residues important for argininosuccinate lyase activity.
Biochemistry, 43, 2004
|
|
1U6V
 
 | | NMR structure of a V3 (IIIB isolate) peptide bound to 447-52D, a human HIV-1 neutralizing antibody | | Descriptor: | V3 peptide | | Authors: | Rosen, O, Chill, J, Sharon, M, Kessler, N, Mester, B, Zolla-Pazner, S, Anglister, J. | | Deposit date: | 2004-08-02 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Induced fit in HIV-neutralizing antibody complexes: evidence for alternative conformations of the gp120 V3 loop and the molecular basis for broad neutralization.
Biochemistry, 44, 2005
|
|
1U4J
 
 | | Crystal structure of a carbohydrate induced dimer of group I phospholipase A2 from Bungarus caeruleus at 2.1 A resolution | | Descriptor: | ACETIC ACID, CHLORIDE ION, SODIUM ION, ... | | Authors: | Singh, G, Gourinath, S, Sharma, S, Bhanumathi, S, Betzel, C, Srinivasan, A, Singh, T.P. | | Deposit date: | 2004-07-26 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of a carbohydrate induced homodimer of phospholipase A(2) from Bungarus caeruleus at 2.1A resolution
J.Struct.Biol., 149, 2005
|
|
1S89
 
 | |
8OF2
 
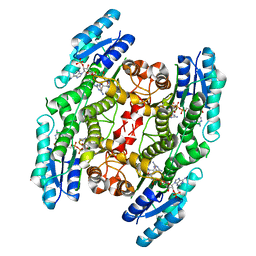 | | Trypanosoma brucei pteridine reductase 1 (TbPTR1) in complex with 2,4,6 triamminopyrimidine (TAP) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tassone, G, Landi, G, Mangani, S, Pozzi, C. | | Deposit date: | 2023-03-13 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The discovery of aryl-2-nitroethyl triamino pyrimidines as anti-Trypanosoma brucei agents.
Eur.J.Med.Chem., 264, 2023
|
|
1S8A
 
 | |
1RQA
 
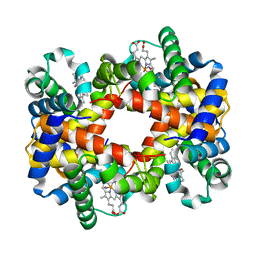 | | Crystallographic Analysis of the Interaction of Nitric Oxide with Quaternary-T Human Hemoglobin. Beta W73E hemoglobin exposed to NO under anaerobic conditions | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, NITRIC OXIDE, ... | | Authors: | Chan, N.-L, Kavanaugh, J.S, Rogers, P.H, Arnone, A. | | Deposit date: | 2003-12-04 | | Release date: | 2005-01-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystallographic analysis of the interaction of nitric oxide with quaternary-T human hemoglobin.
Biochemistry, 43, 2004
|
|
1T8K
 
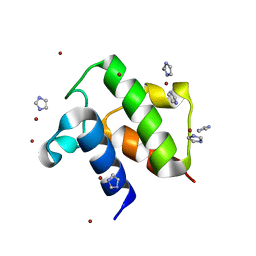 | | Crystal Structure of apo acyl carrier protein from E. coli | | Descriptor: | Acyl carrier protein, IMIDAZOLE, ZINC ION | | Authors: | Qiu, X, Janson, C.A. | | Deposit date: | 2004-05-13 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of apo acyl carrier protein and a proposal to engineer protein crystallization through metal ions.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TFE
 
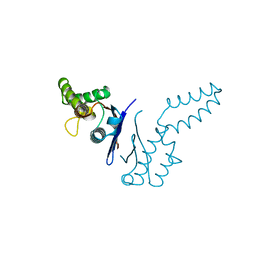 | | DIMERIZATION DOMAIN OF EF-TS FROM T. THERMOPHILUS | | Descriptor: | ELONGATION FACTOR TS | | Authors: | Jiang, Y, Nock, S, Nesper, M, Sprinzl, M, Sigler, P.B. | | Deposit date: | 1996-04-16 | | Release date: | 1996-11-08 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and importance of the dimerization domain in elongation factor Ts from Thermus thermophilus.
Biochemistry, 35, 1996
|
|
1UUD
 
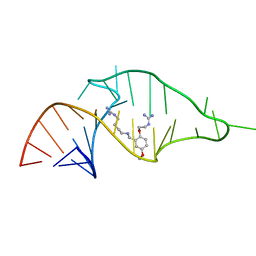 | | NMR structure of a synthetic small molecule, rbt203, bound to HIV-1 TAR RNA | | Descriptor: | N-[2-(2-{[(4-{[AMINO(IMINO)METHYL]AMINO}BUTYL)AMINO]METHYL}-4-METHOXYPHENOXY)ETHYL]GUANIDINE, RNA (5'-(*GP*GP*CP*AP*GP*AP*UP*CP*UP*GP*AP*GP *CP*CP*UP*GP*GP*GP*AP*GP*CP*UP*CP*UP*CP*UP*GP*CP*C) -3') | | Authors: | Davis, B, Afshar, M, Varani, G, Karn, J, Murchie, A.I.H, Lentzen, G, Drysdale, M.J, Potter, A.J, Bower, J, Aboul-Ela, F. | | Deposit date: | 2003-12-18 | | Release date: | 2004-03-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational Design of Inhibitors of HIV-1 Tar RNA Through the Stabilisation of Electrostatic "Hot Spots"
J.Mol.Biol., 336, 2004
|
|
1US8
 
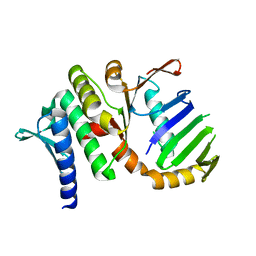 | | The Rad50 signature motif: essential to ATP binding and biological function | | Descriptor: | DNA DOUBLE-STRAND BREAK REPAIR RAD50 ATPASE | | Authors: | Moncalian, G, Lengsfeld, B, Bhaskara, V, Hopfner, K.P, Karcher, A, Alden, E, Tainer, J.A, Paull, T.T. | | Deposit date: | 2003-11-20 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Rad50 Signature Motif: Essential to ATP Binding and Biological Function
J.Mol.Biol., 335, 2004
|
|
1U4C
 
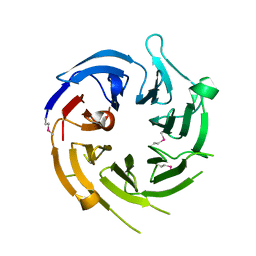 | |
1U6U
 
 | | NMR structure of a V3 (IIIB isolate) peptide bound to 447-52D, a human HIV-1 neutralizing antibody | | Descriptor: | V3 peptide | | Authors: | Rosen, O, Chill, J, Sharon, M, Kessler, N, Mester, B, Zolla-Pazner, S, Anglister, J. | | Deposit date: | 2004-08-02 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Induced fit in HIV-neutralizing antibody complexes: evidence for alternative conformations of the gp120 V3 loop and the molecular basis for broad neutralization.
Biochemistry, 44, 2005
|
|
1RQ4
 
 | | Crystallographic Analysis of the Interaction of Nitric Oxide with Quaternary-T Human Hemoglobin, HEMOGLOBIN EXPOSED TO NO UNDER AEROBIC CONDITIONS | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, NITRIC OXIDE, ... | | Authors: | Chan, N.L, Kavanaugh, J.S, Rogers, P.H, Arnone, A. | | Deposit date: | 2003-12-04 | | Release date: | 2003-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystallographic analysis of the interaction of nitric oxide with quaternary-T human hemoglobin.
Biochemistry, 43, 2004
|
|
1RQ3
 
 | | Crystallographic Analysis of the Interaction of Nitric Oxide with Quaternary-T Human Deoxyhemoglobin, Deoxyhemoglobin | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chan, N.L, Kavanaugh, J.S, Rogers, P.H, Arnone, A. | | Deposit date: | 2003-12-04 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystallographic analysis of the interaction of nitric oxide with quaternary-T human hemoglobin.
Biochemistry, 43, 2004
|
|
1UUI
 
 | | NMR structure of a synthetic small molecule, rbt158, bound to HIV-1 TAR RNA | | Descriptor: | 4-[AMINO(IMINO)METHYL]-1-[2-(3-AMMONIOPROPOXY)-5-METHOXYBENZYL]PIPERAZIN-1-IUM, 5'-R(*GP*GP*CP*AP*GP*AP*UP*CP*UP*GP*AP*GP*CP* CP*UP*GP*GP*GP*AP*GP*CP*UP*CP*UP*CP*UP*GP*CP*C)-3' | | Authors: | Davis, B, Afshar, M, Varani, G, Karn, J, Murchie, A.I.H, Lentzen, G, Drysdale, M.J, Potter, A.J, Bower, J, Aboul-Ela, F. | | Deposit date: | 2003-12-19 | | Release date: | 2004-02-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational Design of Inhibitors of HIV-1 Tar RNA Through the Stabilisation of Electrostatic "Hot Spots"
J.Mol.Biol., 336, 2004
|
|
