1VWQ
 
 | |
2XPX
 
 | | Crystal structure of BHRF1:Bak BH3 complex | | Descriptor: | 1,2-ETHANEDIOL, APOPTOSIS REGULATOR BHRF1, BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, ... | | Authors: | Kvansakul, M, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2010-08-31 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Apoptosis Inhibition by Epstein-Barr Virus Bhrf1.
Plos Pathog., 6, 2010
|
|
1VWO
 
 | |
1N51
 
 | | Aminopeptidase P in complex with the inhibitor apstatin | | Descriptor: | MANGANESE (II) ION, Xaa-Pro aminopeptidase, apstatin | | Authors: | Graham, S.C, Maher, M.J, Lee, M.H, Simmons, W.H, Freeman, H.C, Guss, J.M. | | Deposit date: | 2002-11-03 | | Release date: | 2003-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Escherichia coli aminopeptidase P in complex with the inhibitor apstatin.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1W9A
 
 | | Crystal structure of Rv1155 from Mycobacterium tuberculosis | | Descriptor: | PUTATIVE PYRIDOXINE/PYRIDOXAMINE 5'-PHOSPHATE OXIDASE | | Authors: | Cannan, S, Sulzenbacher, G, Roig-Zamboni, V, Scappuccini, L, Frassinetti, F, Maurien, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2004-10-07 | | Release date: | 2005-01-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Conserved Hypothetical Protein Rv1155 from Mycobacterium Tuberculosis
FEBS Lett., 579, 2005
|
|
1MRQ
 
 | | Crystal structure of human 20alpha-HSD in ternary complex with NADP and 20alpha-hydroxy-progesterone | | Descriptor: | Aldo-keto reductase family 1 member C1, BETA-MERCAPTOETHANOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Couture, J.F, Legrand, P, Cantin, L, Luu-The, V, Labrie, F, Breton, R. | | Deposit date: | 2002-09-18 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Human 20alpha-hydroxysteroid dehydrogenase: crystallographic and site-directed mutagenesis studies lead to the identification of an alternative binding site for C21-steroids.
J.Mol.Biol., 331, 2003
|
|
2XJ7
 
 | | BtGH84 in complex with 6-acetamido-6-deoxy-castanospermine | | Descriptor: | 6-ACETAMIDO-6-DEOXY-CASTANOSPERMINE, CALCIUM ION, O-GLCNACASE BT_4395 | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of O-Glcnacase Using a Potent and Cell-Permeable Inhibitor Does not Induce Insulin Resistance in 3T3-L1 Adipocytes.
Chem.Biol., 17, 2010
|
|
1MTC
 
 | | GLUTATHIONE TRANSFERASE MUTANT Y115F | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, Glutathione S-transferase YB1 | | Authors: | Ladner, J.E, Xiao, G, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 2002-09-20 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Local protein dynamics and catalysis: detection of segmental motion associated with rate-limiting product release by a glutathione transferase
Biochemistry, 41, 2002
|
|
2XLG
 
 | | Structure and metal-loading of a soluble periplasm cupro-protein: Cu- CucA-open | | Descriptor: | COPPER (II) ION, SLL1785 PROTEIN, SODIUM ION | | Authors: | Waldron, K.J, Firbank, S.J, Dainty, S.J, Perez-Rama, M, Tottey, S, Robinson, N.J. | | Deposit date: | 2010-07-20 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Metal Loading of a Soluble Periplasm Cuproprotein.
J.Biol.Chem., 285, 2010
|
|
2XQ0
 
 | | Structure of yeast LTA4 hydrolase in complex with Bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, CHLORIDE ION, LEUKOTRIENE A-4 HYDROLASE, ... | | Authors: | Helgstrand, C, Hasan, M, Usyal, H, Haeggstrom, J.Z, Thunnissen, M.M.G.M. | | Deposit date: | 2010-08-31 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | A Leukotriene A(4) Hydrolase-Related Aminopeptidase from Yeast Undergoes Induced Fit Upon Inhibitor Binding.
J.Mol.Biol., 406, 2011
|
|
2AX3
 
 | |
2AY9
 
 | |
2ALD
 
 | | HUMAN MUSCLE ALDOLASE | | Descriptor: | FRUCTOSE-BISPHOSPHATE ALDOLASE | | Authors: | Dalby, A.R, Littlechild, J.A. | | Deposit date: | 1998-10-21 | | Release date: | 1999-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human muscle aldolase complexed with fructose 1,6-bisphosphate: mechanistic implications.
Protein Sci., 8, 1999
|
|
2AMC
 
 | | Crystal structure of Phenylalanyl-tRNA synthetase complexed with L-tyrosine | | Descriptor: | MAGNESIUM ION, Phenylalanyl-tRNA synthetase alpha chain, Phenylalanyl-tRNA synthetase beta chain, ... | | Authors: | Kotik-Kogan, O, Moor, N, Tworowski, D, Safro, M. | | Deposit date: | 2005-08-09 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Discrimination of L-Phenylalanine from L-Tyrosine by Phenylalanyl-tRNA Synthetase
Structure, 13, 2005
|
|
2AZT
 
 | | Crystal structure of H176N mutant of human Glycine N-Methyltransferase | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Luka, Z, Pakhomova, S, Luka, Y, Newcomer, M.E, Wagner, C. | | Deposit date: | 2005-09-12 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Destabilization of human glycine N-methyltransferase by H176N mutation.
Protein Sci., 16, 2007
|
|
1N5D
 
 | |
2B2K
 
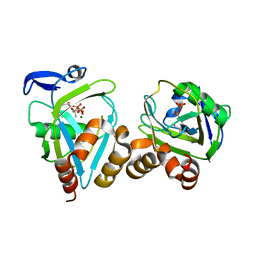 | | structure of Y104F IDI-1 mutant in complex with EIPP | | Descriptor: | 4-HYDROXY-3-METHYL BUTYL DIPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, MAGNESIUM ION, ... | | Authors: | Wouters, J. | | Deposit date: | 2005-09-19 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural role for Tyr-104 in Escherichia coli isopentenyl-diphosphate isomerase: site-directed mutagenesis, enzymology, and protein crystallography
J.Biol.Chem., 281, 2006
|
|
2XP9
 
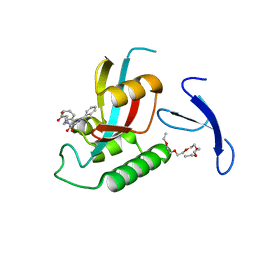 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 4-[BENZYL(CARBOXYMETHYL)CARBAMOYL]-2-PHENYL-1H-IMIDAZOLE-5-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2B3Z
 
 | |
2B5I
 
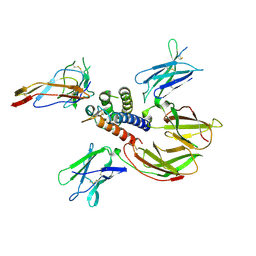 | | cytokine receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common gamma chain, ... | | Authors: | Wang, X, Rickert, M, Garcia, K.C. | | Deposit date: | 2005-09-28 | | Release date: | 2005-11-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors.
Science, 310, 2005
|
|
2AW1
 
 | | Carbonic anhydrase inhibitors: Valdecoxib binds to a different active site region of the human isoform II as compared to the structurally related cyclooxygenase II "selective" inhibitor Celecoxib | | Descriptor: | 4-(5-METHYL-3-PHENYLISOXAZOL-4-YL)BENZENESULFONAMIDE, 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase II, ... | | Authors: | Di Fiore, A, Pedone, C, D'Ambrosio, K, Scozzafava, A, De Simone, G, Supuran, C.T. | | Deposit date: | 2005-08-31 | | Release date: | 2006-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Carbonic anhydrase inhibitors: Valdecoxib binds to a different active site region of the human isoform II as compared to the structurally related cyclooxygenase II
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1MPM
 
 | | MALTOPORIN MALTOSE COMPLEX | | Descriptor: | MAGNESIUM ION, MALTOPORIN, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Dutzler, R, Schirmer, T. | | Deposit date: | 1996-01-11 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of various maltooligosaccharides bound to maltoporin reveal a specific sugar translocation pathway.
Structure, 4, 1996
|
|
1TAT
 
 | |
2Y2W
 
 | | Elucidation of the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. | | Descriptor: | ARABINOFURANOSIDASE | | Authors: | Lagaert, S, Schoepe, J, Delcour, J.A, Lavigne, R, Strelkov, S.V, Courtin, C.M, Mikkelsen, N.E, Sandgren, M, Volckaert, G. | | Deposit date: | 2010-12-16 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Elucidation of the Substrate Specificity and Protein Structure of Abfb, a Family 51 Alpha-L- Arabinofuranosidase from Bifidobacterium Longum.
To be Published
|
|
1LW3
 
 | | Crystal Structure of Myotubularin-related protein 2 complexed with phosphate | | Descriptor: | Myotubularin-related protein 2, PHOSPHATE ION | | Authors: | Begley, M.J, Taylor, G.S, Kim, S.-A, Veine, D.M, Dixon, J.E, Stuckey, J.A. | | Deposit date: | 2002-05-30 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Phosphoinositide Phosphatase, MTMR2: Insights into Myotubular Myopathy and Charcot-Marie-Tooth Syndrome
Mol.Cell, 12, 2003
|
|
