1TCD
 
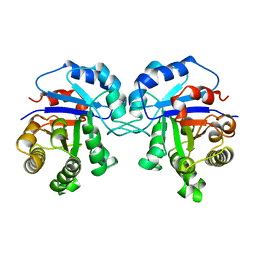 | | TRYPANOSOMA CRUZI TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Maldonado, E, Soriano-Garcia, M, Cabrera, N, Garza-Ramos, G, Tuena De Gomez-Puyou, M, Gomez-Puyou, A, Perez-Montfort, R. | | Deposit date: | 1998-01-29 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Differences in the intersubunit contacts in triosephosphate isomerase from two closely related pathogenic trypanosomes.
J.Mol.Biol., 283, 1998
|
|
1TCE
 
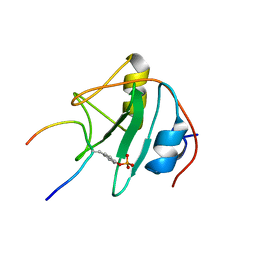 | | SOLUTION NMR STRUCTURE OF THE SHC SH2 DOMAIN COMPLEXED WITH A TYROSINE-PHOSPHORYLATED PEPTIDE FROM THE T-CELL RECEPTOR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PHOSPHOPEPTIDE OF THE ZETA CHAIN OF T CELL RECEPTOR, SHC | | Authors: | Zhou, M.-M, Meadows, R.P, Logan, T.M, Yoon, H.S, Wade, W.R, Ravichandran, K.S, Burakoff, S.J, Feisk, S.W. | | Deposit date: | 1996-03-27 | | Release date: | 1997-05-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Shc SH2 domain complexed with a tyrosine-phosphorylated peptide from the T-cell receptor.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1TCF
 
 | |
1TCG
 
 | |
1TCH
 
 | |
1TCJ
 
 | |
1TCK
 
 | |
1TCM
 
 | |
1TCO
 
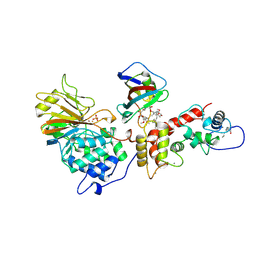 | | TERNARY COMPLEX OF A CALCINEURIN A FRAGMENT, CALCINEURIN B, FKBP12 AND THE IMMUNOSUPPRESSANT DRUG FK506 (TACROLIMUS) | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, FE (III) ION, ... | | Authors: | Griffith, J.P, Kim, J.L, Kim, E.E, Sintchak, M.D, Thomson, J.A, Fitzgibbon, M.J, Fleming, M.A, Caron, P.R, Hsiao, K, Navia, M.A. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex.
Cell(Cambridge,Mass.), 82, 1995
|
|
1TCP
 
 | |
1TCR
 
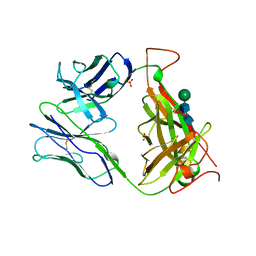 | | MURINE T-CELL ANTIGEN RECEPTOR 2C CLONE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Garcia, K.C, Degano, M, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 1996-09-12 | | Release date: | 1997-03-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex.
Science, 274, 1996
|
|
1TCS
 
 | |
1TCU
 
 | | Crystal Structure of the Purine Nucleoside Phosphorylase from Schistosoma mansoni in complex with phosphate and acetate | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Pereira, H.D, Franco, G.R, Cleasby, A, Garratt, R.C. | | Deposit date: | 2004-05-21 | | Release date: | 2005-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures for the Potential Drug Target Purine Nucleoside Phosphorylase from Schistosoma mansoni Causal Agent of Schistosomiasis.
J.Mol.Biol., 353, 2005
|
|
1TCV
 
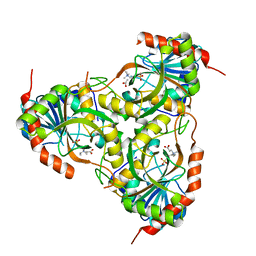 | | Crystal Structure of the Purine Nucleoside Phosphorylase from Schistosoma mansoni in complex with Non-detergent Sulfobetaine 195 and acetate | | Descriptor: | ACETATE ION, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, purine-nucleoside phosphorylase | | Authors: | Pereira, H.D, Franco, G.R, Cleasby, A, Garratt, R.C. | | Deposit date: | 2004-05-21 | | Release date: | 2005-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures for the Potential Drug Target Purine Nucleoside Phosphorylase from Schistosoma mansoni Causal Agent of Schistosomiasis.
J.Mol.Biol., 353, 2005
|
|
1TCW
 
 | | SIV PROTEASE COMPLEXED WITH INHIBITOR SB203386 | | Descriptor: | (2R,4S,5S,1'S)-2-PHENYLMETHYL-4-HYDROXY-5-(TERT-BUTOXYCARBONYL)AMINO-6-PHENYL HEXANOYL-N-(1'-IMIDAZO-2-YL)-2'-METHYLPROPANAMIDE, SIV PROTEASE | | Authors: | Hoog, S.S, Abdel-Meguid, S.S. | | Deposit date: | 1996-06-05 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human immunodeficiency virus protease ligand specificity conferred by residues outside of the active site cavity.
Biochemistry, 35, 1996
|
|
1TCX
 
 | | HIV TRIPLE MUTANT PROTEASE COMPLEXED WITH INHIBITOR SB203386 | | Descriptor: | (2R,4S,5S,1'S)-2-PHENYLMETHYL-4-HYDROXY-5-(TERT-BUTOXYCARBONYL)AMINO-6-PHENYL HEXANOYL-N-(1'-IMIDAZO-2-YL)-2'-METHYLPROPANAMIDE, HIV PROTEASE | | Authors: | Hoog, S.S, Abdel-Meguid, S.S. | | Deposit date: | 1996-06-05 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human immunodeficiency virus protease ligand specificity conferred by residues outside of the active site cavity.
Biochemistry, 35, 1996
|
|
1TCY
 
 | |
1TCZ
 
 | |
1TD0
 
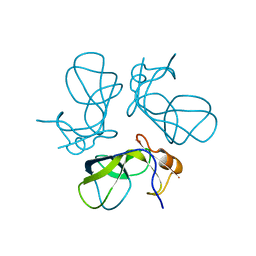 | | Viral capsid protein SHP at pH 5.5 | | Descriptor: | Head decoration protein | | Authors: | Chang, C, Forrer, P, Ott, D, Wlodawer, A, Plueckthun, A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Kinetic Stability and Crystal Structure of the Viral Capsid Protein SHP
J.Mol.Biol., 344, 2004
|
|
1TD1
 
 | | Crystal Structure of the Purine Nucleoside Phosphorylase from Schistosoma mansoni in complex with acetate | | Descriptor: | ACETATE ION, purine-nucleoside phosphorylase | | Authors: | Pereira, H.D, Franco, G.R, Cleasby, A, Garratt, R.C. | | Deposit date: | 2004-05-21 | | Release date: | 2005-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures for the Potential Drug Target Purine Nucleoside Phosphorylase from Schistosoma mansoni Causal Agent of Schistosomiasis.
J.Mol.Biol., 353, 2005
|
|
1TD2
 
 | | Crystal Structure of the PdxY Protein from Escherichia coli | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, Pyridoxamine kinase, SULFATE ION | | Authors: | Safo, M.K, Musayev, F.N, Hunt, S, di Salvo, M, Scarsdale, N, Schirch, V. | | Deposit date: | 2004-05-21 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of the PdxY Protein from Escherichia coli
J.Bacteriol., 186, 2004
|
|
1TD3
 
 | | Crystal structure of VSHP_BPP21 in space group C2 | | Descriptor: | Head decoration protein | | Authors: | Chang, C, Forrer, P, Ott, D, Wlodawer, A, Plueckthun, A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Kinetic Stability and Crystal Structure of the Viral Capsid Protein SHP.
J.Mol.Biol., 344, 2004
|
|
1TD4
 
 | | Crystal structure of VSHP_BPP21 in space group H3 with high resolution. | | Descriptor: | Head decoration protein | | Authors: | Chang, C, Forrer, P, Ott, D, Wlodawer, A, Plueckthun, A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-11-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kinetic Stability and Crystal Structure of the Viral Capsid Protein SHP.
J.Mol.Biol., 344, 2004
|
|
1TD5
 
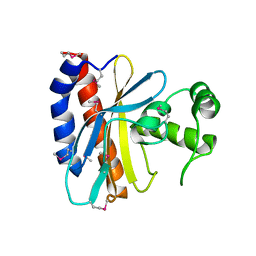 | | Crystal Structure of the Ligand Binding Domain of E. coli IclR. | | Descriptor: | Acetate operon repressor | | Authors: | Walker, J.R, Evdokimova, L, Zhang, R.-G, Bochkarev, A, Joachimiak, A, Arrowsmith, C, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-21 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analyses of the Ligand Binding Sites of the IclR family of transcriptional regulators
To be Published
|
|
1TD6
 
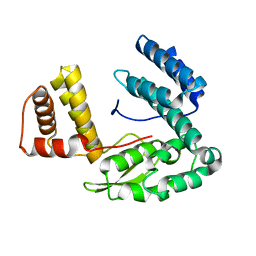 | | Crystal structure of the conserved hypothetical protein MP506/MPN330 (gi: 1674200)from Mycoplasma pneumoniae | | Descriptor: | Hypothetical protein MG237 homolog | | Authors: | Das, D, Oganesyan, N, Yokota, H, Jancarik, J, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the conserved hypothetical protein MPN330 (GI: 1674200) from Mycoplasma pneumoniae.
Proteins, 58, 2004
|
|
