8T8Q
 
 | | Identification of GDC-1971 (RLY-1971), a SHP2 inhibitor designed for the treatment of solid tumors | | Descriptor: | 1-[(3P)-3-(3-chloro-2-fluorophenyl)-1H-pyrazolo[3,4-b]pyrazin-6-yl]-4-methylpiperidin-4-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Tang, Y, Nguyen, V, Wilbur, J.D. | | Deposit date: | 2023-06-23 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Identification of GDC-1971 (RLY-1971), a SHP2 Inhibitor Designed for the Treatment of Solid Tumors.
J.Med.Chem., 66, 2023
|
|
6R0I
 
 | | Glycogen Phosphorylase b in complex with 4 | | Descriptor: | Glycogen phosphorylase, muscle form, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Koulas, M.S, Tsagkarakou, S.A, Kyriakis, E, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-17 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High Consistency of Structure-Based Design and X-Ray Crystallography: Design, Synthesis, Kinetic Evaluation and Crystallographic Binding Mode Determination of Biphenyl-N-acyl-beta-d-Glucopyranosylamines as Glycogen Phosphorylase Inhibitors.
Molecules, 24, 2019
|
|
8SWU
 
 | |
6Z2F
 
 | |
6I5N
 
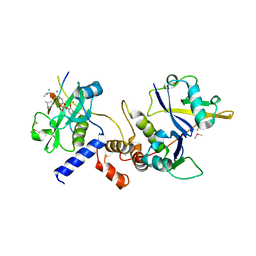 | | Crystal structure of SOCS2:Elongin C:Elongin B in complex with growth hormone receptor peptide | | Descriptor: | COBALT (II) ION, Elongin-B, Elongin-C, ... | | Authors: | Kung, W.W, Ramachandran, S, Makukhin, N, Bruno, E, Ciulli, A. | | Deposit date: | 2018-11-14 | | Release date: | 2019-05-29 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural insights into substrate recognition by the SOCS2 E3 ubiquitin ligase.
Nat Commun, 10, 2019
|
|
6DF7
 
 | |
8BB7
 
 | |
5H87
 
 | | Crystal structure of mRojoA mutant - P63H - W143S | | Descriptor: | mRojoA fluorescent protein | | Authors: | Pandelieva, A.T, Tremblay, V, Sarvan, S, Chica, R.A, Couture, J.-F. | | Deposit date: | 2015-12-23 | | Release date: | 2016-01-27 | | Last modified: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Brighter Red Fluorescent Proteins by Rational Design of Triple-Decker Motif.
Acs Chem.Biol., 11, 2016
|
|
6I6S
 
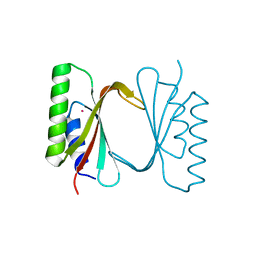 | | Circular permutant of ribosomal protein S6, adding 9aa to C terminal of P68-69, L75A mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6, POTASSIUM ION, SODIUM ION | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Exposing the distinctive modular behavior of beta-strands and alpha-helices in folded proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7PGH
 
 | | NaVAe1/Sp1CTDp (DDM) | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DODECAETHYLENE GLYCOL, DODECANE, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-29 | | Method: | X-RAY DIFFRACTION (4.194 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5HH6
 
 | |
7K6Z
 
 | | Carbonic Anhydrase IX mimic complexed with 4-(2-(3-(4-fluorophenyl)ureido)ethylsulfonamido)benzenesulfonamide | | Descriptor: | 4-{[(2-{[(4-fluorophenyl)carbamoyl]amino}ethyl)sulfonyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Andring, J.T, Singh, S, McKenna, R. | | Deposit date: | 2020-09-21 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.657 Å) | | Cite: | Handling drug-target selectivity: A study on ureido containing Carbonic Anhydrase inhibitors.
Eur.J.Med.Chem., 212, 2021
|
|
7PGG
 
 | | NaVAb1p detergent (DM) | | Descriptor: | 2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}ethyl heptadecanoate, Ion transport protein | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6D9T
 
 | | BshA from Staphylococcus aureus complexed with UDP | | Descriptor: | Glycosyl transferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Royer, C.R, Cook, P.D. | | Deposit date: | 2018-04-30 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and functional analysis of the glycosyltransferase BshA from Staphylococcus aureus: Insights into the reaction mechanism and regulation of bacillithiol production.
Protein Sci., 28, 2019
|
|
8BRT
 
 | | Crystal structure of a variant of penicillin G acylase from Bacillaceae i. s. sp. FJAT-27231 with reduced surface entropy and additionally engineered crystal contact | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wichmann, J, Mayer, J, Mattes, H, Lukat, P, Blankenfeldt, W, Biedendieck, R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Multistep Engineering of a Penicillin G Acylase for Systematic Improvement of Crystallization Efficiency
Cryst.Growth Des., 2023
|
|
5HHF
 
 | |
6I7J
 
 | | Crystal structure of monomeric FICD mutant L258D | | Descriptor: | Adenosine monophosphate-protein transferase FICD, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
7Q0T
 
 | | Lysozyme soaked with V(IV)OSO4 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Santos, M.F.A, Fernandes, A.C.P, Correia, I, Sciortino, G, Garribba, E, Santos-Silva, T, Pessoa, J.C. | | Deposit date: | 2021-10-16 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Binding of V IV O 2+ , V IV OL, V IV OL 2 and V V O 2 L Moieties to Proteins: X-ray/Theoretical Characterization and Biological Implications.
Chemistry, 28, 2022
|
|
6OQO
 
 | | CDK6 in complex with Cpd24 N-(5-(6-ethyl-2,6-diazaspiro[3.3]heptan-2-yl)pyridin-2-yl)-5-fluoro-4-(4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl)pyrimidin-2-amine | | Descriptor: | Cyclin-dependent kinase 6, N-[5-(6-ethyl-2,6-diazaspiro[3.3]heptan-2-yl)pyridin-2-yl]-5-fluoro-4-[(4R)-4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl]pyrimidin-2-amine | | Authors: | Murray, J.M, Boenig, G.D.L. | | Deposit date: | 2019-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6I74
 
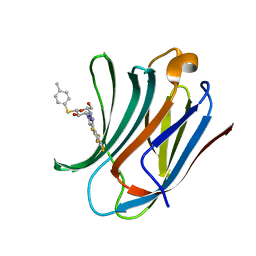 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative 1 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-4-[4-[2,3,4,5,6-pentakis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.959 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
5VS2
 
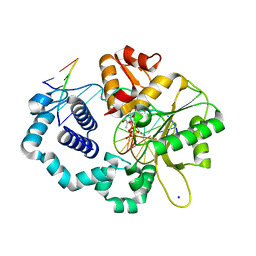 | |
7K7W
 
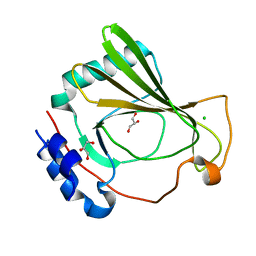 | | The X-ray crystal structure of SSR4, an S. pombe chromatin remodelling protein: native | | Descriptor: | CHLORIDE ION, GLYCEROL, SWI/SNF and RSC complexes subunit ssr4 | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2020-09-24 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The X-ray crystal structure of the N-terminal domain of Ssr4, a Schizosaccharomyces pombe chromatin-remodelling protein.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5HIF
 
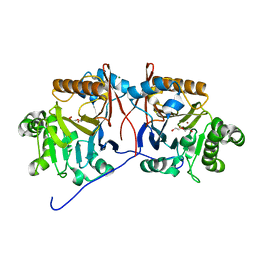 | | Crystal structure of a reconstructed lactonase ancestor, Anc1-MPH, of the bacterial methyl parathion hydrolase, MPH. | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ZINC ION, ... | | Authors: | Baier, F, Carr, P.D, Jackson, C.J. | | Deposit date: | 2016-01-11 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | to be published
To Be Published
|
|
6Z61
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-[6-azanyl-8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-[6-(2-azanylethylamino)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy]prop-1-ynyl]purin-9-yl]-5-(hydroxymethyl)oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2020-05-27 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | New Chemical Probe Targeting Bacterial NAD Kinase.
Molecules, 25, 2020
|
|
5HIP
 
 | |
