6NO0
 
 | |
6WT2
 
 | | Crystal Structure of Putative NAD(P)H-Flavin Oxidoreductase from Neisseria meningitidis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Crofts, T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-01 | | Release date: | 2020-05-13 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional and Structural Characterization of Diverse NfsB Chloramphenicol Reductase Enzymes from Human Pathogens.
Microbiol Spectr, 10, 2022
|
|
5C66
 
 | |
7KL1
 
 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and GluN2B(S1303D) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Calcium/calmodulin-dependent protein kinase type II subunit alpha, ... | | Authors: | Ozden, C, Stratton, M.M, Garman, S.C. | | Deposit date: | 2020-10-28 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
5CCX
 
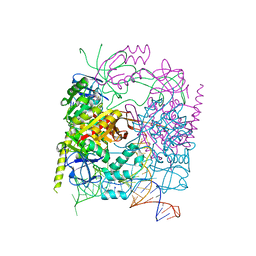 | | Structure of the product complex of tRNA m1A58 methyltransferase with tRNA3Lys as substrate | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A, ... | | Authors: | Finer-Moore, J, Czudnochowski, N, O'Connell III, J.D, Wang, A.L, Stroud, R.M. | | Deposit date: | 2015-07-02 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human tRNA m(1)A58 Methyltransferase-tRNA3(Lys) Complex: Refolding of Substrate tRNA Allows Access to the Methylation Target.
J.Mol.Biol., 427, 2015
|
|
7KL0
 
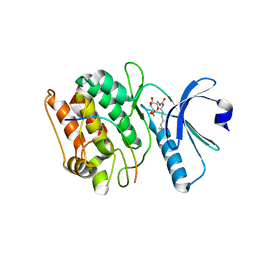 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and GluN2B(S1303D) | | Descriptor: | 1,2-ETHANEDIOL, Calcium/calmodulin-dependent protein kinase type II subunit alpha, Glutamate receptor ionotropic, ... | | Authors: | Ozden, C, Stratton, M.M, Garman, S.C. | | Deposit date: | 2020-10-28 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
5CCB
 
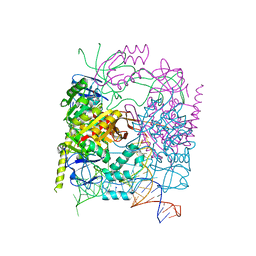 | | Crystal structure of human m1A58 methyltransferase in a complex with tRNA3Lys and SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A, ... | | Authors: | Finer-Moore, J, Czudnochowski, N, O'Connell III, J.D, Wang, A.L, Stroud, R.M. | | Deposit date: | 2015-07-01 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human tRNA m(1)A58 Methyltransferase-tRNA3(Lys) Complex: Refolding of Substrate tRNA Allows Access to the Methylation Target.
J.Mol.Biol., 427, 2015
|
|
7L5M
 
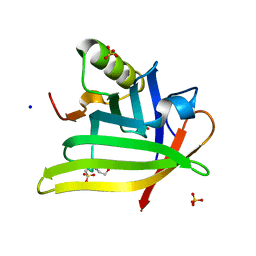 | | Crystal Structure of the DiB-RM-split Protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lipocalin family protein, SODIUM ION, ... | | Authors: | Bozhanova, N.G, Harp, J.M, Meiler, J. | | Deposit date: | 2020-12-22 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Computational redesign of a fluorogen activating protein with Rosetta.
Plos Comput.Biol., 17, 2021
|
|
7L5L
 
 | |
7L5K
 
 | | Crystal structure of the DiB-RM protein | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, ISOPROPYL ALCOHOL, Lipocalin family protein, ... | | Authors: | Bozhanova, N.G, Harp, J.M, Meiler, J. | | Deposit date: | 2020-12-22 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Computational redesign of a fluorogen activating protein with Rosetta.
Plos Comput.Biol., 17, 2021
|
|
7L4X
 
 | |
7L4Y
 
 | |
7JRQ
 
 | | Crystallographically Characterized De Novo Designed Mn-Diphenylporphyrin Binding Protein | | Descriptor: | CHLORIDE ION, GLYCEROL, MPP1, ... | | Authors: | Mann, S.I, DeGrado, W.F. | | Deposit date: | 2020-08-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | De Novo Design, Solution Characterization, and Crystallographic Structure of an Abiological Mn-Porphyrin-Binding Protein Capable of Stabilizing a Mn(V) Species.
J.Am.Chem.Soc., 143, 2021
|
|
5A7B
 
 | |
7OVG
 
 | |
7K9X
 
 | |
7K14
 
 | | Ternary soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN and methanesulfonate | | Descriptor: | Alkanesulfonate monooxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liew, J.J.M, Dowling, D.P, El Saudi, I.M. | | Deposit date: | 2020-09-07 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7K64
 
 | | Binary titrated soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN | | Descriptor: | Alkanesulfonate monooxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liew, J.J.M, Dowling, D.P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7K9L
 
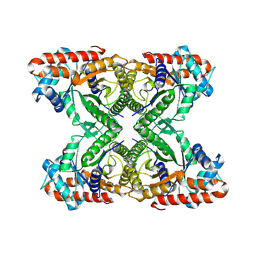 | | Aldolase, rabbit muscle (no beam-tilt refinement) | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Cianfrocco, M.A, Kearns, S.E, Cash, J.N, Li, Y. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | High-resolution cryo-EM using beam-image shift at 200 keV.
Iucrj, 7, 2020
|
|
7KA2
 
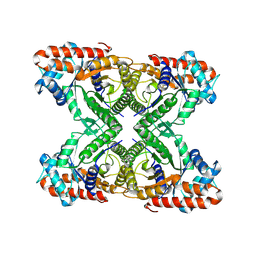 | | Aldolase, rabbit muscle (beam-tilt refinement x2) | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Kearns, S.K, Cash, J.N, Cianfrocco, M.A, Li, Y. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution cryo-EM using beam-image shift at 200 keV.
Iucrj, 7, 2020
|
|
7L9X
 
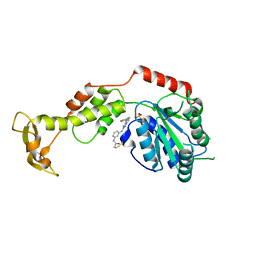 | | Structure of VPS4B in complex with an allele-specific covalent inhibitor | | Descriptor: | N-{3-[(8-phenyl[1,2,4]triazolo[1,5-a]pyridin-2-yl)amino]phenyl}propanamide, SULFATE ION, Vacuolar protein sorting-associated protein 4B | | Authors: | Grasso, M, Cupido, T, Kapoor, T.M. | | Deposit date: | 2021-01-05 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A chemical genetics approach to examine the functions of AAA proteins.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KA3
 
 | | Aldolase, rabbit muscle (beam-tilt refinement x3) | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Kearns, S.K, Cash, J.N, Cianfrocco, M.A, Li, Y. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | High-resolution cryo-EM using beam-image shift at 200 keV.
Iucrj, 7, 2020
|
|
7KA4
 
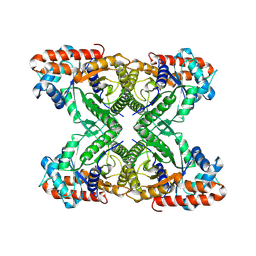 | | Aldolase, rabbit muscle (beam-tilt refinement x4) | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Kearns, S.K, Cash, J.N, Cianfrocco, M.A, Li, Y. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | High-resolution cryo-EM using beam-image shift at 200 keV.
Iucrj, 7, 2020
|
|
5CD1
 
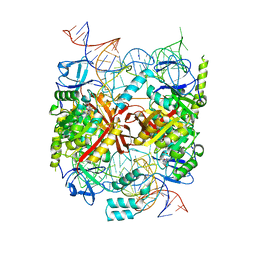 | | Structure of an asymmetric tetramer of human tRNA m1A58 methyltransferase in a complex with SAH and tRNA3Lys | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A, ... | | Authors: | Finer-Moore, J, Czudnochowski, N, O'Connell III, J.D, Wang, A.L, Stroud, R.M. | | Deposit date: | 2015-07-02 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal Structure of the Human tRNA m(1)A58 Methyltransferase-tRNA3(Lys) Complex: Refolding of Substrate tRNA Allows Access to the Methylation Target.
J.Mol.Biol., 427, 2015
|
|
7RTH
 
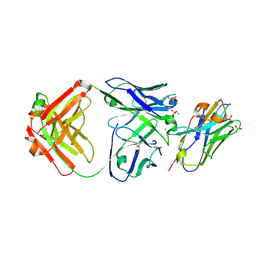 | | Crystal structure of an anti-lysozyme nanobody in complex with an anti-nanobody Fab "NabFab" | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Fragment Antigen-Binding Heavy Chain, ... | | Authors: | Filippova, E.V, Mukherjee, S, Bloch, J.S, Locher, K.P, Kossiakoff, A.A. | | Deposit date: | 2021-08-13 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
