4IOQ
 
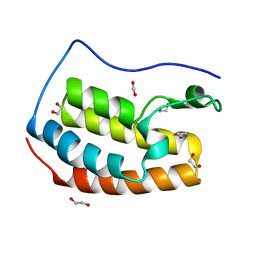 | |
1WE7
 
 | | Solution structure of Ubiquitin-like domain in SF3a120 | | Descriptor: | Sf3a1 protein | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-24 | | Release date: | 2004-11-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Ubiquitin-like domain in SF3a120
To be Published
|
|
4IIR
 
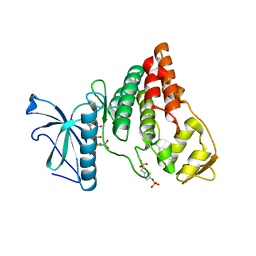 | | Crystal Structure of AMPPNP-bound Human PRPF4B kinase domain | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Mechin, I, Haas, K, Chen, X, Zhang, Y, McLean, L. | | Deposit date: | 2012-12-20 | | Release date: | 2013-08-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evaluation of Cancer Dependence and Druggability of PRP4 Kinase Using Cellular, Biochemical, and Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
4IOR
 
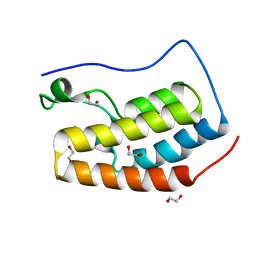 | |
1WM2
 
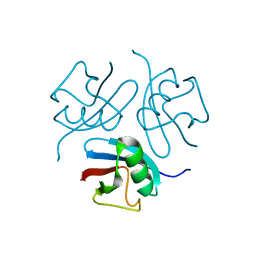 | |
1WTB
 
 | | Complex structure of the C-terminal RNA-binding domain of hnRNP D (AUF1) with telomere DNA | | Descriptor: | 5'-D(P*TP*AP*GP*G)-3', Heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Enokizono, Y, Konishi, Y, Nagata, K, Ouhashi, K, Uesugi, S, Ishikawa, F, Katahira, M. | | Deposit date: | 2004-11-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D
J.Biol.Chem., 280, 2005
|
|
4J0R
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(R)-hydroxy(phenyl)methyl]phenol, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Hewings, D.S, von Delft, F, Conway, S.J, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Optimization of 3,5-dimethylisoxazole derivatives as potent bromodomain ligands.
J.Med.Chem., 56, 2013
|
|
1WF0
 
 | | Solution structure of RRM domain in TAR DNA-binding protein-43 | | Descriptor: | TAR DNA-binding protein-43 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in TAR DNA-binding protein-43
To be Published
|
|
1WIB
 
 | | Solution structure of the N-terminal domain from mouse hypothetical protein BAB22488 | | Descriptor: | 60S ribosomal protein L12 | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain from mouse hypothetical protein BAB22488
To be Published
|
|
1WIZ
 
 | | Solution structure of the first CUT domain of KIAA1034 protein | | Descriptor: | DNA-binding protein SATB2 | | Authors: | Inoue, K, Nameki, S, Hayashi, F, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first CUT domain of KIAA1034 protein
To be Published
|
|
4J3I
 
 | | X-ray crystal structure of bromodomain complex to 1.24 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain-containing protein 4, ... | | Authors: | Stein, A.J, White, A, Suto, R.K. | | Deposit date: | 2013-02-05 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | RVX-208, an Inducer of ApoA-I in Humans, Is a BET Bromodomain Antagonist.
Plos One, 8, 2013
|
|
1WHW
 
 | | Solution structure of the N-terminal RNA binding domain from hypothetical protein BAB23448 | | Descriptor: | hypothetical protein RIKEN CDNA 1200009A02 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal RNA binding domain from hypothetical protein BAB23448
To be Published
|
|
1X0F
 
 | | Complex structure of the C-terminal RNA-binding domain of hnRNP D(AUF1) with telomeric DNA | | Descriptor: | 5'-D(P*TP*AP*GP*G)-3', Heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Enokizono, Y, Konishi, Y, Nagata, K, Ouhashi, K, Uesugi, S, Ishikawa, F, Katahira, M. | | Deposit date: | 2005-03-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D.
J.Biol.Chem., 280, 2005
|
|
1X4A
 
 | | Solution structure of RRM domain in splicing factor SF2 | | Descriptor: | splicing factor, arginine/serine-rich 1 (splicing factor 2, alternate splicing factor) variant | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in splicing factor SF2
To be Published
|
|
4JD2
 
 | |
4JBO
 
 | | Novel Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules | | Descriptor: | 1-(4-{2-[(6-{4-[2-(dimethylamino)ethoxy]phenyl}furo[2,3-d]pyrimidin-4-yl)amino]ethyl}phenyl)-3-phenylurea, Aurora kinase A | | Authors: | Wu, J.S, Leou, J.S, Peng, Y.H, Hsueh, C.C, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2013-02-20 | | Release date: | 2013-06-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JNK
 
 | | Lactate Dehydrogenase A in complex with inhibitor compound 22 | | Descriptor: | (2R)-2-{[5-cyano-4-(3,4-dichlorophenyl)-6-oxo-1,6-dihydropyrimidin-2-yl]sulfanyl}-N-(4-sulfamoylphenyl)propanamide, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Identification of substituted 2-thio-6-oxo-1,6-dihydropyrimidines as inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1XB7
 
 | | X-ray structure of ERRalpha LBD in complex with a PGC-1alpha peptide at 2.5A resolution | | Descriptor: | IODIDE ION, Peroxisome proliferator activated receptor gamma coactivator 1 alpha, Steroid hormone receptor ERR1 | | Authors: | Kallen, J, Schlaeppi, J.M, Bitsch, F, Filipuzzi, I, Schilb, A, Riou, V, Graham, A, Strauss, A, Geiser, M, Fournier, B. | | Deposit date: | 2004-08-30 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence for Ligand-independent Transcriptional Activation of the Human Estrogen-related Receptor {alpha} (ERR{alpha}): CRYSTAL STRUCTURE OF ERR{alpha} LIGAND BINDING DOMAIN IN COMPLEX WITH PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR COACTIVATOR-1{alpha}
J.Biol.Chem., 279, 2004
|
|
1XKT
 
 | | Human fatty acid synthase: Structure and substrate selectivity of the thioesterase domain | | Descriptor: | fatty acid synthase | | Authors: | Chakravarty, B, Gu, Z, Chirala, S.S, Wakil, S.J, Quiocho, F.A. | | Deposit date: | 2004-09-29 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human fatty acid synthase: structure and substrate selectivity of the thioesterase domain.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1XG1
 
 | | Solution structure of Myb-domain of human TRF2 | | Descriptor: | Telomeric repeat binding factor 2 | | Authors: | Paquet, F, Meudal, H, Amiard, S, Doudeau, M, Paoletti, J, Giraud-Panis, M.J, Lancelot, G. | | Deposit date: | 2004-09-16 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR studies of telomeric nucleoprotein complexes involving the Myb-like domain of the human telomeric protein TRF2
C.R.Chimie, 9, 2006
|
|
4KMD
 
 | | Crystal structure of Sufud60-Gli1p | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Sufu, ... | | Authors: | Zhang, Y, Qi, X, Zhang, Z, Wu, G. | | Deposit date: | 2013-05-08 | | Release date: | 2013-11-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci.
Nat Commun, 4, 2013
|
|
4KPW
 
 | |
1W9G
 
 | | Structure of ERH (Enhencer of Rudimentary Gene) | | Descriptor: | ENHANCER OF RUDIMENTARY HOMOLOG | | Authors: | Wan, C, Tempel, W, Liu, Z, Wang, B.-C, Rose, R.B. | | Deposit date: | 2004-10-13 | | Release date: | 2005-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Conserved Transcriptional Repressor Enhancer of Rudimentary Homolog
Biochemistry, 44, 2005
|
|
1W8L
 
 | |
4KLN
 
 | | Structure of p97 N-D1 A232E mutant in complex with ATPgS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Xia, D, Tang, W.K. | | Deposit date: | 2013-05-07 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Altered Intersubunit Communication Is the Molecular Basis for Functional Defects of Pathogenic p97 Mutants.
J.Biol.Chem., 288, 2013
|
|
