3PF3
 
 | | Crystal structure of a mutant (C202A) of Triosephosphate isomerase from Giardia lamblia derivatized with MMTS | | Descriptor: | CALCIUM ION, GLYCEROL, SULFATE ION, ... | | Authors: | Enriquez-Flores, S, Rodriguez-Romero, A, Hernandez-Santoyo, A, Reyes-Vivas, H. | | Deposit date: | 2010-10-27 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Determining the molecular mechanism of inactivation by chemical modification of triosephosphate isomerase from the human parasite Giardia lamblia: A study for antiparasitic drug design.
Proteins, 79, 2011
|
|
7AYG
 
 | | oxalyl-CoA decarboxylase from Methylorubrum extorquens with bound TPP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative oxalyl-CoA decarboxylase (Oxc, ... | | Authors: | Pfister, P, Burgener, S, Nattermann, M, Zarzycki, J, Erb, T.J. | | Deposit date: | 2020-11-12 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering a Highly Efficient Carboligase for Synthetic One-Carbon Metabolism.
Acs Catalysis, 11, 2021
|
|
7B2E
 
 | | quadruple mutant of oxalyl-CoA decarboxylase from Methylorubrum extorquens with bound TPP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative oxalyl-CoA decarboxylase (Oxc, ... | | Authors: | Pfister, P, Burgener, S, Nattermann, M, Zarzycki, J, Erb, T.J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Engineering a Highly Efficient Carboligase for Synthetic One-Carbon Metabolism.
Acs Catalysis, 11, 2021
|
|
3PKP
 
 | | Q83S Variant of S. Enterica RmlA with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Glucose-1-phosphate thymidylyltransferase, MAGNESIUM ION | | Authors: | Chang, A, Moretti, R, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Expanding the Nucleotide and Sugar 1-Phosphate Promiscuity of Nucleotidyltransferase RmlA via Directed Evolution.
J.Biol.Chem., 286, 2011
|
|
4S2U
 
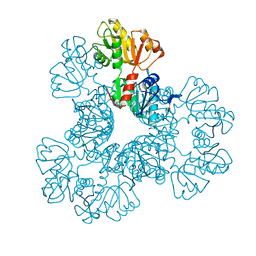 | | Crystal structure of the Phosphorybosylpyrophosphate synthetase from E. Coli | | Descriptor: | MAGNESIUM ION, Ribose-phosphate pyrophosphokinase | | Authors: | Timofeev, V.I, Abramchik, Y.A, Muravieva, T.I, Iaroslavtceva, A.K, Stepanenko, V.N, Zhukhlistova, N.E, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2015-01-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of the Phosphorybosylpyrophosphate synthetase from E. Coli
To be Published
|
|
6WBO
 
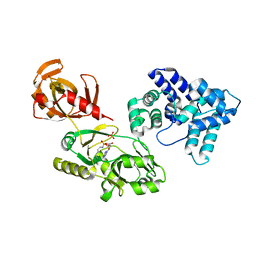 | |
7SX5
 
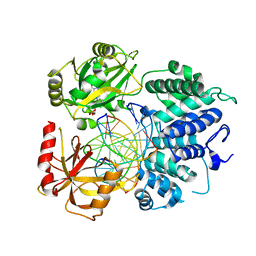 | | Crystal structure of ligase I with nick duplexes containing mismatch A:C | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA chain 1, DNA chain 2, ... | | Authors: | Tang, Q, Gulkis, M, McKenna, R, Caglayan, M. | | Deposit date: | 2021-11-22 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of LIG1 that engage with mutagenic mismatches inserted by pol beta in base excision repair.
Nat Commun, 13, 2022
|
|
7SXE
 
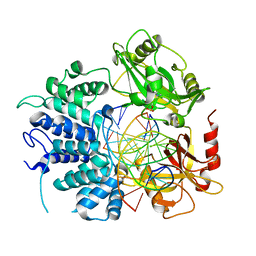 | | Crystal structure of ligase I with nick duplexes containing cognate G:T | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA chain 1, DNA chain 2, ... | | Authors: | Tang, Q, Gulkis, M, McKenna, R, Caglayan, M. | | Deposit date: | 2021-11-22 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of LIG1 that engage with mutagenic mismatches inserted by pol beta in base excision repair.
Nat Commun, 13, 2022
|
|
7ZUB
 
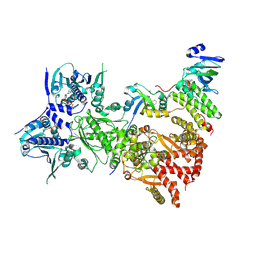 | | Cryo-EM structure of the indirubin-bound Hsp90-XAP2-AHR complex | | Descriptor: | (3~{Z})-3-(3-oxidanylidene-1~{H}-indol-2-ylidene)-1~{H}-indol-2-one, ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, ... | | Authors: | Gruszczyk, J, Savva, C.G, Lai-Kee-Him, J, Bous, J, Ancelin, A, Kwong, H.S, Grandvuillemin, L, Bourguet, W. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of the agonist-bound Hsp90-XAP2-AHR cytosolic complex.
Nat Commun, 13, 2022
|
|
5KCV
 
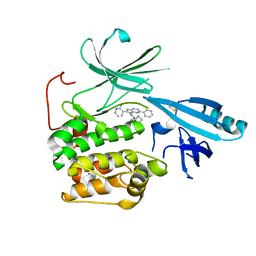 | | Crystal structure of allosteric inhibitor, ARQ 092, in complex with autoinhibited form of AKT1 | | Descriptor: | 3-[3-[4-(1-azanylcyclobutyl)phenyl]-5-phenyl-imidazo[4,5-b]pyridin-2-yl]pyridin-2-amine, RAC-alpha serine/threonine-protein kinase | | Authors: | Eathiraj, S. | | Deposit date: | 2016-06-07 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of 3-(3-(4-(1-Aminocyclobutyl)phenyl)-5-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amine (ARQ 092): An Orally Bioavailable, Selective, and Potent Allosteric AKT Inhibitor.
J.Med.Chem., 59, 2016
|
|
4QVH
 
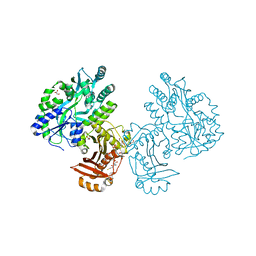 | | Crystal structure of the essential Mycobacterium tuberculosis phosphopantetheinyl transferase PptT, solved as a fusion protein with maltose binding protein | | Descriptor: | CITRATE ANION, COENZYME A, GLYCEROL, ... | | Authors: | Jung, J, Bashiri, G, Johnston, J.M, Baker, E.N. | | Deposit date: | 2014-07-15 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the essential Mycobacterium tuberculosis phosphopantetheinyl transferase PptT, solved as a fusion protein with maltose binding protein.
J.Struct.Biol., 188, 2014
|
|
6P0E
 
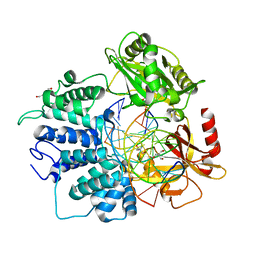 | | Human DNA Ligase 1 (E346A,E592A) bound to adenylated DNA containing an 8-oxo guanine:adenine base-pair | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schellenberg, M.J, Williams, R.S, Tumbale, P.S, Riccio, A.A. | | Deposit date: | 2019-05-16 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Two-tiered enforcement of high-fidelity DNA ligation.
Nat Commun, 10, 2019
|
|
5KPX
 
 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure IV) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
5KHA
 
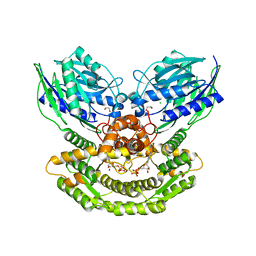 | |
7ALE
 
 | | Crystal structure of human PAICS in complex with inhibitor 69 | | Descriptor: | (2~{S})-2-[[5-azanyl-1-[(2~{R},3~{R},4~{S},5~{R})-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]imidazol-4-yl]car bonylamino]butanedioic acid, 2-azanyl-~{N}-[2-bromanyl-5-[4-[3-(dimethylamino)propylsulfonyl]piperazin-1-yl]phenyl]-1,3-oxazole-4-carboxamide, Multifunctional protein ADE2 | | Authors: | Skerlova, J, Marttila, P, Unterlass, J, Jemth, A.-S, Henriksson, M, Wakchaure, P, Grube, M, Warpman Berglund, U, Homan, E, Helleday, T, Stenmark, P. | | Deposit date: | 2020-10-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Cellular and biochemical validation of a potent PAICS inhibitor
To Be Published
|
|
3MNQ
 
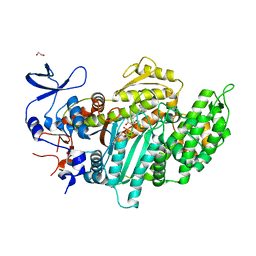 | | Crystal structure of myosin-2 motor domain in complex with ADP-metavanadate and resveratrol | | Descriptor: | 1,2-ETHANEDIOL, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Schneider, J, Taft, M, Backhaus, A, Baruch, P, Fedorov, R, Manstein, D.J. | | Deposit date: | 2010-04-22 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of resveratrol regulation of myosin activity.
To be Published
|
|
4QSL
 
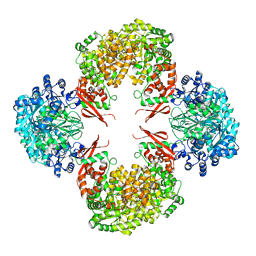 | |
8VZM
 
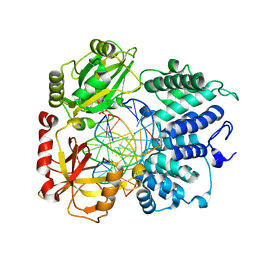 | | DNA Ligase 1 captured with pre-step 3 ligation at the rA:T nicksite | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*GP*AP*C)-3'), ... | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2024-02-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
8VDT
 
 | | DNA Ligase 1 with nick DNA 3'rA:T | | Descriptor: | DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*TP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA ligase 1, DNA/RNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*T)-R(P*A)-D(P*GP*TP*CP*GP*GP*AP*C)-3'), ... | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2023-12-17 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
8VZL
 
 | | DNA Ligase 1 captured with pre-step 3 ligation at the rG:C nicksite | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*GP*AP*C)-3'), ... | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2024-02-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
8VDS
 
 | | DNA Ligase 1 with nick DNA 3'rG:C | | Descriptor: | DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA ligase 1, DNA/RNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*T)-R(P*G)-D(P*GP*TP*CP*GP*GP*AP*C)-3') | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2023-12-17 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
8VDN
 
 | | DNA Ligase 1 with nick dG:C | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase 1, Downstream Oligo, ... | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2023-12-16 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
8HZ5
 
 | |
6VR7
 
 | | Structure of a pseudomurein peptide ligase type C from Methanothermus fervidus | | Descriptor: | ACETOACETIC ACID, GLYCEROL, Mur ligase middle domain protein, ... | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Archaeal pseudomurein and bacterial murein cell wall biosynthesis share a common evolutionary ancestry
FEMS Microbes, 2, 2021
|
|
5CSL
 
 | |
