8HPJ
 
 | |
8H7B
 
 | | The crystal structure of human mcl1 kinase domain in complex with MCL1-M-EBA | | Descriptor: | 7-[3-(isoquinolin-7-yloxymethyl)-1,5-dimethyl-pyrazol-4-yl]-3-(3-naphthalen-1-yloxypropyl)-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhu, C.J, Zhang, Z.M. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.46408451 Å) | | Cite: | 2-Ethynylbenzaldehyde-Based, Lysine-Targeting Irreversible Covalent Inhibitors for Protein Kinases and Nonkinases.
J.Am.Chem.Soc., 2023
|
|
2DSV
 
 | | Interactions of protective signalling factor with chitin-like polysaccharide: Crystal structure of the complex between signalling protein from sheep (SPS-40) and a hexasaccharide at 2.5A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-4)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Kumar, J, Singh, N, Sharma, S, Bhushan, A, Srinivasan, A, Singh, T.P. | | Deposit date: | 2006-07-07 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Carbohydrate binding properties and carbohydrate induced conformational switch in sheep secretory glycoprotein (SPS-40): crystal structures of four complexes of SPS-40 with chitin-like oligosaccharides
J.Struct.Biol., 158, 2007
|
|
8GZ1
 
 | | Cryo-EM structure of human NaV1.6/beta1/beta2,apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, Sodium channel protein type 8 subunit alpha, ... | | Authors: | Li, Y, Jiang, D. | | Deposit date: | 2022-09-24 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human Na V 1.6 channel reveals Na + selectivity and pore blockade by 4,9-anhydro-tetrodotoxin.
Nat Commun, 14, 2023
|
|
1JTS
 
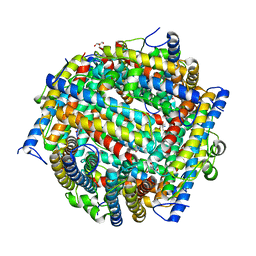 | |
8GZ2
 
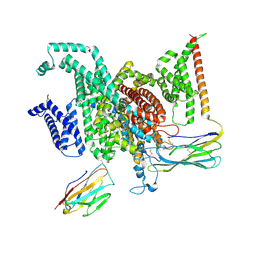 | | Cryo-EM structure of human NaV1.6/beta1/beta2-4,9-anhydro-tetrodotoxin | | Descriptor: | (1R,2S,3S,4R,5R,9S,11S,12S,14R)-7-amino-2,4,12-trihydroxy-2-(hydroxymethyl)-10,13,15-trioxa-6,8-diazapentacyclo[7.4.1.1~3,12~.0~5,11~.0~5,14~]pentadec-7-en-8-ium (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 8 subunit alpha, ... | | Authors: | Li, Y, Jiang, D. | | Deposit date: | 2022-09-24 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human Na V 1.6 channel reveals Na + selectivity and pore blockade by 4,9-anhydro-tetrodotoxin.
Nat Commun, 14, 2023
|
|
2DTU
 
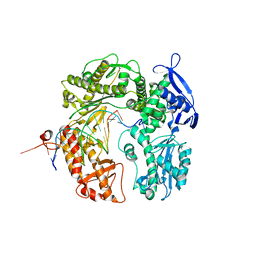 | | Crystal structure of the beta hairpin loop deletion variant of RB69 gp43 in complex with DNA containing an abasic site analog | | Descriptor: | 5'-D(*CP*GP*(3DR)P*CP*TP*TP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*CP*G)-3', 5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*AP*AP*GP*A)-3', DNA polymerase | | Authors: | Aller, P, Hogg, M, Konigsberg, W, Wallace, S.S, Doublie, S. | | Deposit date: | 2006-07-15 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural and biochemical investigation of the role in proofreading of a beta hairpin loop found in the exonuclease domain of a replicative DNA polymerase of the B family.
J.Biol.Chem., 282, 2007
|
|
2DFC
 
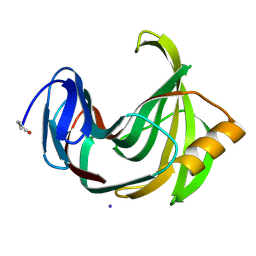 | | Xylanase II from Tricoderma reesei at 293K | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Harata, K, Akiba, T. | | Deposit date: | 2006-02-28 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structure of an orthorhombic form of xylanase II from Trichoderma reesei and analysis of thermal displacement.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2DCK
 
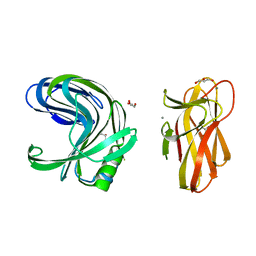 | | A tetragonal-lattice structure of alkaliphilic XynJ from Bacillus sp. 41M-1 | | Descriptor: | CALCIUM ION, GLYCEROL, xylanase J | | Authors: | Fibriansah, G, Ihsanawati, Tanaka, N, Nakamura, S, Kumasaka, T. | | Deposit date: | 2006-01-07 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A two-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1
To be Published
|
|
2DH9
 
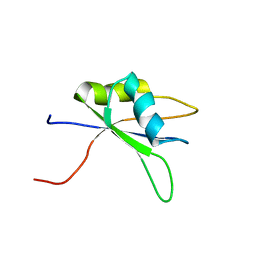 | | Solution structure of the C-terminal RNA binding domain in Heterogeneous nuclear ribonucleoprotein M | | Descriptor: | Heterogeneous nuclear ribonucleoprotein M | | Authors: | Ochi, T, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-23 | | Release date: | 2006-09-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal RNA binding domain in Heterogeneous nuclear ribonucleoprotein M
To be Published
|
|
1JYN
 
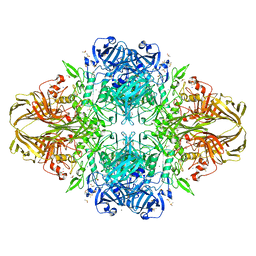 | |
2DDK
 
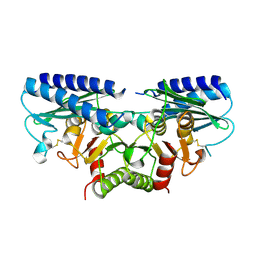 | | Crystal structure of human myo-inositol monophosphatase 2 (IMPA2) (orthorhombic form) | | Descriptor: | Inositol monophosphatase 2 | | Authors: | Arai, R, Ito, K, Kamo-Uchikubo, T, Bessho, Y, Ohba, H, Ohnishi, T, Yoshikawa, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-30 | | Release date: | 2007-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human myo-inositol monophosphatase 2, the product of the putative susceptibility gene for bipolar disorder, schizophrenia, and febrile seizures
Proteins, 67, 2007
|
|
1JMQ
 
 | | YAP65 (L30K mutant) WW domain in Complex with GTPPPPYTVG peptide | | Descriptor: | 65 KDA YES-ASSOCIATED PROTEIN, WW Domain Binding Protein-1 | | Authors: | Pires, J.R, Taha-Nejad, F, Toepert, F, Ast, T, Hoffmuller, U, Schneider-Mergener, J, Kuhne, R, Macias, M.J, Oschkinat, H. | | Deposit date: | 2001-07-19 | | Release date: | 2001-12-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the YAP65 WW domain and the variant L30 K in complex with the peptides GTPPPPYTVG, N-(n-octyl)-GPPPY and PLPPY and the application of peptide libraries reveal a minimal binding epitope.
J.Mol.Biol., 314, 2001
|
|
2DN1
 
 | | 1.25A resolution crystal structure of human hemoglobin in the oxy form | | Descriptor: | Hemoglobin alpha subunit, Hemoglobin beta subunit, OXYGEN MOLECULE, ... | | Authors: | Park, S.-Y, Yokoyama, T, Shibayama, N, Shiro, Y, Tame, J.R. | | Deposit date: | 2006-04-25 | | Release date: | 2006-05-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25 a resolution crystal structures of human haemoglobin in the oxy, deoxy and carbonmonoxy forms.
J.Mol.Biol., 360, 2006
|
|
1JP3
 
 | | Structure of E.coli undecaprenyl pyrophosphate synthase | | Descriptor: | 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, undecaprenyl pyrophosphate synthase | | Authors: | Ko, T.P, Chen, Y.K, Robinson, H, Tsai, P.C, Gao, Y.G, Chen, A.P.C, Wang, A.H.J, Liang, P.H. | | Deposit date: | 2001-07-31 | | Release date: | 2001-08-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli undecaprenyl-pyrophosphate synthase catalysis.
J.Biol.Chem., 276, 2001
|
|
2DIN
 
 | | Solution structure of the Myb_DNA-binding domain of human Cell division cycle 5-like protein | | Descriptor: | Cell division cycle 5-like protein | | Authors: | Yoneyama, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-30 | | Release date: | 2007-04-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Myb_DNA-binding domain of human Cell division cycle 5-like protein
To be Published
|
|
1JI1
 
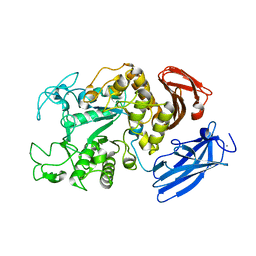 | | Crystal Structure Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase 1 | | Descriptor: | ALPHA-AMYLASE I, CALCIUM ION | | Authors: | Kamitori, S, Abe, A, Ohtaki, A, Kaji, A, Tonozuka, T, Sakano, Y. | | Deposit date: | 2001-06-28 | | Release date: | 2002-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures and structural comparison of Thermoactinomyces vulgaris R-47 alpha-amylase 1 (TVAI) at 1.6 A resolution and alpha-amylase 2 (TVAII) at 2.3 A resolution.
J.Mol.Biol., 318, 2002
|
|
1JRG
 
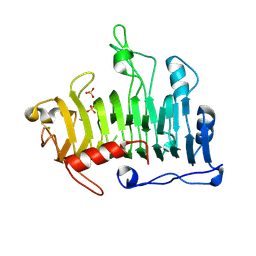 | | Crystal Structure of the R3 form of Pectate Lyase A, Erwinia chrysanthemi | | Descriptor: | Pectate lyase, SULFATE ION | | Authors: | Thomas, L.M, Doan, C, Oliver, R.L, Yoder, M.D. | | Deposit date: | 2001-08-13 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of pectate lyase A: comparison to other isoforms.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1JKH
 
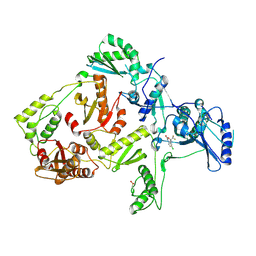 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DMP-266(EFAVIRENZ) | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-12 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLG
 
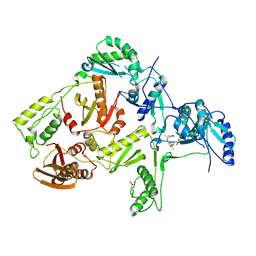 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JOF
 
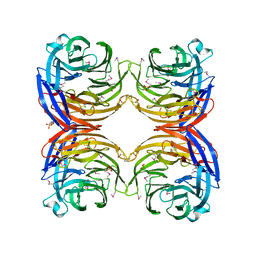 | | Neurospora crassa 3-carboxy-cis,cis-mucoante lactonizing enzyme | | Descriptor: | BETA-MERCAPTOETHANOL, CARBOXY-CIS,CIS-MUCONATE CYCLASE, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), ... | | Authors: | Kajander, T, Merckel, M.C, Thompson, A, Deacon, A.M, Mazur, P, Kozarich, J.W, Goldman, A. | | Deposit date: | 2001-07-28 | | Release date: | 2002-04-12 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of Neurospora crassa 3-carboxy-cis,cis-muconate lactonizing enzyme, a beta propeller cycloisomerase.
Structure, 10, 2002
|
|
1JU3
 
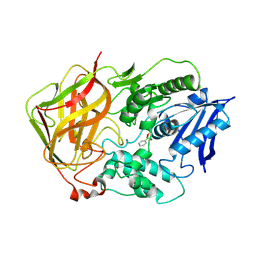 | | BACTERIAL COCAINE ESTERASE COMPLEX WITH TRANSITION STATE ANALOG | | Descriptor: | PHENYL BORONIC ACID, cocaine esterase | | Authors: | Larsen, N.A, Turner, J.M, Stevens, J, Rosser, S.J, Basran, A, Lerner, R.A, Bruce, N.C, Wilson, I.A. | | Deposit date: | 2001-08-23 | | Release date: | 2001-12-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of a bacterial cocaine esterase.
Nat.Struct.Biol., 9, 2002
|
|
1JR8
 
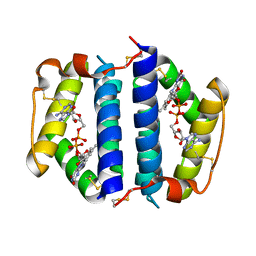 | | Crystal Structure of Erv2p | | Descriptor: | Erv2 PROTEIN, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Gross, E, Sevier, C.S, Vala, A, Kaiser, C.A, Fass, D. | | Deposit date: | 2001-08-13 | | Release date: | 2001-12-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A new FAD-binding fold and intersubunit disulfide shuttle in the thiol oxidase Erv2p.
Nat.Struct.Biol., 9, 2002
|
|
8HHG
 
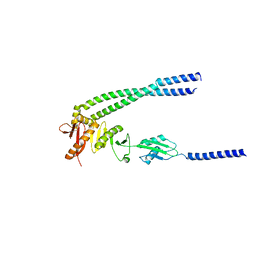 | | The bacterial divisome protein complex FtsB-FtsL-FtsQ | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the heterotrimeric membrane protein complex FtsB-FtsL-FtsQ of the bacterial divisome.
Nat Commun, 14, 2023
|
|
8HHH
 
 | | The bacterial divisome protein complex FtsB-FtsL-FtsQ | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the heterotrimeric membrane protein complex FtsB-FtsL-FtsQ of the bacterial divisome.
Nat Commun, 14, 2023
|
|
