6F8Y
 
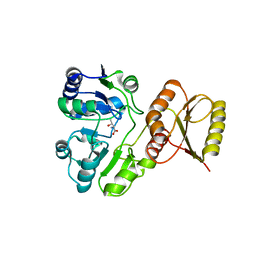 | | Crystal structure of P. abyssi Sua5 complexed with L-threonine | | Descriptor: | THREONINE, Threonylcarbamoyl-AMP synthase | | Authors: | Pichard-Kostuch, A, Zhang, W, Liger, D, Daugeron, M.C, Letoquart, J, Li de la Sierra-Gallay, I, Forterre, P, Collinet, B, van Tilbeurgh, H, Basta, T. | | Deposit date: | 2017-12-13 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure-function analysis of Sua5 protein reveals novel functional motifs required for the biosynthesis of the universal t6A tRNA modification.
RNA, 24, 2018
|
|
5WS2
 
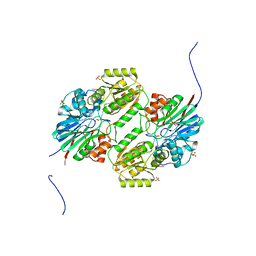 | | Crystal structure of mpy-RNase J (mutant S247A), an archaeal RNase J from Methanolobus psychrophilus R15, complex with RNA | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*A)-3'), Ribonuclease J, SULFATE ION, ... | | Authors: | Li, D.F, Feng, N. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | New molecular insights into an archaeal RNase J reveal a conserved processive exoribonucleolysis mechanism of the RNase J family
Mol. Microbiol., 106, 2017
|
|
4REO
 
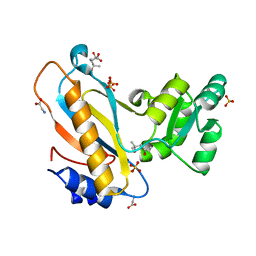 | | Mutant ribosomal protein l1 from thermus thermophilus with threonine 217 replaced by valine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 50S ribosomal protein L1, GLYCINE, ... | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, Tishchenko, S.V, Nikonov, S.V. | | Deposit date: | 2014-09-23 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7OVS
 
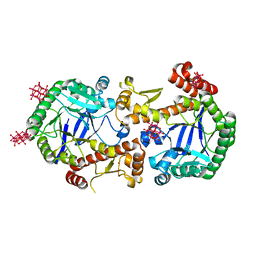 | | Heterodimeric murine tRNA-guanine transglycosylase in the presence of Anderson-Evans type (TEW) and Strandberg type polyoxometalate (POM) | | Descriptor: | 6-tungstotellurate(VI), Queuine tRNA-ribosyltransferase accessory subunit 2, Queuine tRNA-ribosyltransferase catalytic subunit 1, ... | | Authors: | Sebastiani, M, Heine, A, Reuter, K. | | Deposit date: | 2021-06-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Biochemical Investigation of the Heterodimeric Murine tRNA-Guanine Transglycosylase.
Acs Chem.Biol., 17, 2022
|
|
7OWZ
 
 | | Heterodimeric murine tRNA-guanine transglycosylase in complex with queuine and in the presence of Anderson-Evans type (TEW) and Strandberg type polyoxometalate (POM) | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, 6-tungstotellurate(VI), Queuine tRNA-ribosyltransferase accessory subunit 2, ... | | Authors: | Sebastiani, M, Heine, A, Reuter, K. | | Deposit date: | 2021-06-21 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Biochemical Investigation of the Heterodimeric Murine tRNA-Guanine Transglycosylase.
Acs Chem.Biol., 17, 2022
|
|
8UK6
 
 | |
7KX9
 
 | | Cryo-EM structure of Ephydatia fluviatilis PiwiA-piRNA-target complex | | Descriptor: | MAGNESIUM ION, Piwi-A, RNA (5'-R(P*UP*CP*UP*CP*UP*UP*GP*AP*GP*UP*UP*GP*GP*AP*CP*AP*AP*AP*UP*GP*GP*CP*AP*(OMG))-3'), ... | | Authors: | Anzelon, T.A, Chowdhury, S, Lander, G.C, MacRae, I.J. | | Deposit date: | 2020-12-03 | | Release date: | 2021-07-14 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for piRNA targeting.
Nature, 597, 2021
|
|
4PUO
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with RNA/DNA and Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', 5'-R(P*AP*UP*GP*GP*UP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*UP*GP*UP*G)-3', ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-03-13 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
5LSL
 
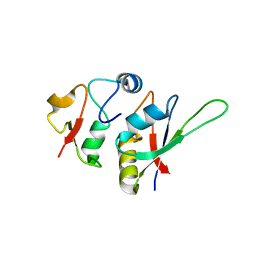 | | Crystal structure of yeast Hsh49p in complex with Cus1p binding domain. | | Descriptor: | Cold sensitive U2 snRNA suppressor 1, Protein HSH49 | | Authors: | van Roon, A.M, Obayashi, E, Sposito, B, Oubridge, C, Nagai, K. | | Deposit date: | 2016-09-02 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of U2 snRNP SF3b components: Hsh49p in complex with Cus1p-binding domain.
RNA, 23, 2017
|
|
7D5C
 
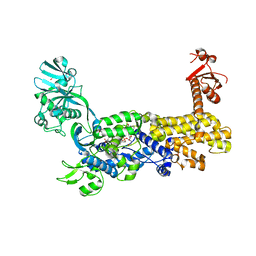 | | IleRS in complex with a tRNA site inhibitor | | Descriptor: | (2E,4S,5S,6E,8E)-10-[(2S,3R,6S,8R,9S)-3-butyl-9-methyl-2-[(1E,3E)-3-methyl-5-oxidanyl-5-oxidanylidene-penta-1,3-dienyl]-3-(4-oxidanyl-4-oxidanylidene-butanoyl)oxy-1,7-dioxaspiro[5.5]undecan-8-yl]-4,8-dimethyl-5-oxidanyl-deca-2,6,8-trienoic acid, 1,2-ETHANEDIOL, Isoleucine--tRNA ligase, ... | | Authors: | Chen, B, Luo, S, Zhou, H. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Inhibitory mechanism of reveromycin A at the tRNA binding site of a class I synthetase.
Nat Commun, 12, 2021
|
|
7KX7
 
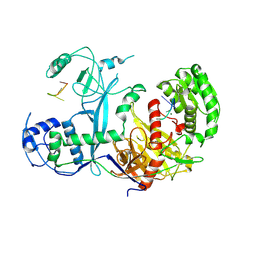 | | Cryo-EM structure of Ephydatia fluviatilis PiwiA-piRNA complex | | Descriptor: | MAGNESIUM ION, Piwi-A, RNA (5'-R(P*UP*CP*UP*CP*AP*GP*(OMC))-3') | | Authors: | Anzelon, T.A, Chowdhury, S, Lander, G.C, MacRae, I.J. | | Deposit date: | 2020-12-03 | | Release date: | 2021-07-14 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for piRNA targeting.
Nature, 597, 2021
|
|
1P6V
 
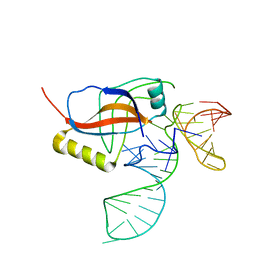 | | Crystal structure of the tRNA domain of transfer-messenger RNA in complex with SmpB | | Descriptor: | 45-MER, SsrA-binding protein | | Authors: | Gutmann, S, Haebel, P.W, Metzinger, L, Sutter, M, Felden, B, Ban, N. | | Deposit date: | 2003-04-30 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB
Nature, 424, 2003
|
|
5ONA
 
 | | Drosophila Bag-of-marbles CBM peptide bound to human CAF40-CNOT1 | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 9, CHLORIDE ION, ... | | Authors: | Raisch, T, Sgromo, A, Backhaus, C, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2017-08-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | DrosophilaBag-of-marbles directly interacts with the CAF40 subunit of the CCR4-NOT complex to elicit repression of mRNA targets.
RNA, 24, 2018
|
|
5ONB
 
 | | Drosophila Bag-of-marbles CBM peptide bound to human CAF40 | | Descriptor: | CCR4-NOT transcription complex subunit 9, GLYCEROL, Protein bag-of-marbles | | Authors: | Raisch, T, Sgromo, A, Backhaus, C, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2017-08-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DrosophilaBag-of-marbles directly interacts with the CAF40 subunit of the CCR4-NOT complex to elicit repression of mRNA targets.
RNA, 24, 2018
|
|
8OWX
 
 | | Crystal Structure of METTL6 bound to SAH | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA N(3)-methylcytidine methyltransferase METTL6 | | Authors: | Throll, P, Basu, S, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis of tRNA recognition by the m 3 C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8OWY
 
 | | Crystal structure of METTL6 mutant 40-269 bound to SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA N(3)-methylcytidine methyltransferase METTL6 | | Authors: | Throll, P, Basu, S, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of tRNA recognition by the m 3 C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5TEF
 
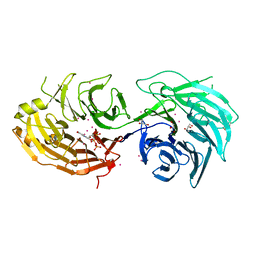 | | Crystal structure of Gemin5 WD40 repeats in complex with m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, GLYCEROL, Gem-associated protein 5, ... | | Authors: | Chao, X, Tempel, W, Bian, C, He, H, Cerovina, T, Seitova, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-21 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly.
Genes Dev., 30, 2016
|
|
5U9B
 
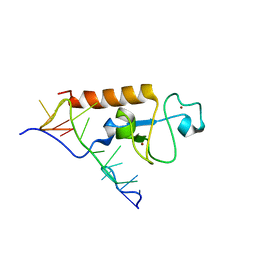 | | Solution structure of the zinc fingers 1 and 2 of MBNL1 in complex with human cardiac troponin T pre-mRNA | | Descriptor: | Muscleblind-like protein 1, RNA (5'-R(P*GP*UP*CP*UP*CP*GP*CP*UP*UP*UP*UP*CP*CP*CP*C)-3'), ZINC ION | | Authors: | Phukan, P.D, Park, S, Martinez-Yamout, M.M, Zeeb, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-12-15 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Interaction of the Tandem Zinc Finger Domains of Human Muscleblind with Cognate RNA from Human Cardiac Troponin T.
Biochemistry, 56, 2017
|
|
2DER
 
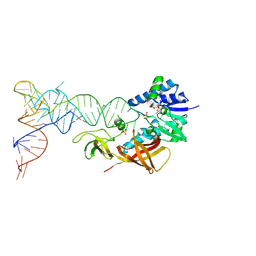 | | Cocrystal structure of an RNA sulfuration enzyme MnmA and tRNA-Glu in the initial tRNA binding state | | Descriptor: | PHOSPHATE ION, SULFATE ION, tRNA, ... | | Authors: | Numata, T, Ikeuchi, Y, Fukai, S, Suzuki, T, Nureki, O. | | Deposit date: | 2006-02-17 | | Release date: | 2006-08-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Snapshots of tRNA sulphuration via an adenylated intermediate
Nature, 442, 2006
|
|
2DET
 
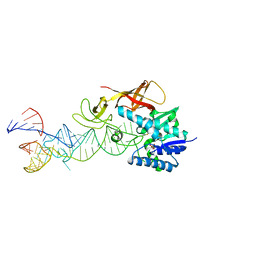 | | Cocrystal structure of an RNA sulfuration enzyme MnmA and tRNA-Glu in the pre-reaction state | | Descriptor: | SULFATE ION, tRNA, tRNA-specific 2-thiouridylase mnmA | | Authors: | Numata, T, Ikeuchi, Y, Fukai, S, Suzuki, T, Nureki, O. | | Deposit date: | 2006-02-17 | | Release date: | 2006-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Snapshots of tRNA sulphuration via an adenylated intermediate
Nature, 442, 2006
|
|
2F4V
 
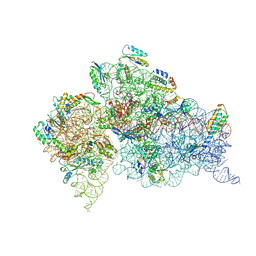 | | 30S ribosome + designer antibiotic | | Descriptor: | (2R)-4-AMINO-N-{(1R,2S,3R,4R,5S)-5-AMINO-2-{2-[(2-AMINOETHYL)AMINO]ETHOXY}-4-[(2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-3-HYDROXYCYCLOHEXYL}-2-HYDROXYBUTANAMIDE, (2S,4S,4AR,5AS,6S,11R,11AS,12R,12AR)-7-CHLORO-4-(DIMETHYLAMINO)-6,10,11,12-TETRAHYDROXY-1,3-DIOXO-1,2,3,4,4A,5,5A,6,11,11A,12,12A-DODECAHYDROTETRACENE-2-CARBOXAMIDE, 16S ribosomal RNA, ... | | Authors: | Murray, J.B, Meroueh, S.O, Russell, R.J, Lentzen, G, Haddad, J, Mobashery, S. | | Deposit date: | 2005-11-24 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Interactions of designer antibiotics and the bacterial ribosome aminoacyl-tRNA site
Chem.Biol., 13, 2006
|
|
2DEU
 
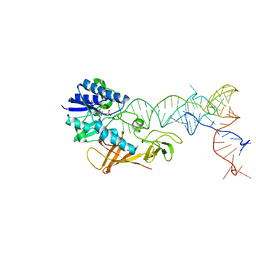 | | Cocrystal structure of an RNA sulfuration enzyme MnmA and tRNA-Glu in the adenylated intermediate state | | Descriptor: | ADENOSINE MONOPHOSPHATE, SULFATE ION, tRNA, ... | | Authors: | Numata, T, Ikeuchi, Y, Fukai, S, Suzuki, T, Nureki, O. | | Deposit date: | 2006-02-17 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Snapshots of tRNA sulphuration via an adenylated intermediate
Nature, 442, 2006
|
|
4G25
 
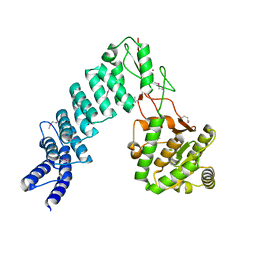 | | Crystal Structure of proteinaceous RNase P 1 (PRORP1) from A. thaliana, SeMet substituted form with Sr | | Descriptor: | Pentatricopeptide repeat-containing protein At2g32230, mitochondrial, STRONTIUM ION, ... | | Authors: | Koutmos, M, Howard, M.J, Fierke, C.A. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mitochondrial ribonuclease P structure provides insight into the evolution of catalytic strategies for precursor-tRNA 5' processing.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4G23
 
 | |
4FVU
 
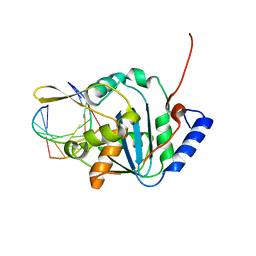 | | Structural basis for the dsRNA specificity of the Lassa virus NP exonuclease | | Descriptor: | MAGNESIUM ION, Nucleoprotein, RNA (5'-R(*CP*UP*CP*CP*CP*UP*CP*C)-3'), ... | | Authors: | Hastie, K.M, King, L.B, Zandonatti, M.A, Saphire, E.O. | | Deposit date: | 2012-06-29 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural Basis for the dsRNA Specificity of the Lassa Virus NP Exonuclease.
Plos One, 7, 2012
|
|
