8PP2
 
 | |
8PP0
 
 | | Crystal structure of Retinoic Acid Receptor alpha (RXRA) in complexed with JP147 | | Descriptor: | 3-[4-[2,3-dihydro-1H-inden-4-yl(methyl)amino]-6-(trifluoromethyl)pyrimidin-2-yl]oxypropanoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chaikuad, A, Pollinger, J, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-07-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of a Highly Potent Partial RXR Agonist with Superior Physicochemical Properties.
J.Med.Chem., 67, 2024
|
|
8POW
 
 | | Crystal Structure of the C19G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the air-oxidized state at 1.61 A Resolution. | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, Fe4S4, ... | | Authors: | Kalms, J, Schmidt, A, Scheerer, P. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
8POV
 
 | | Crystal Structure of the C19G/C120G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the H2-reduced state at 1.92 A Resolution. | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Schmidt, A, Kalms, J, Scheerer, P. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
8POU
 
 | | Crystal Structure of the C19G/C120G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the air-oxidized state at 1.65 A Resolution. | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Schmidt, A, Kalms, J, Scheerer, P. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
8POT
 
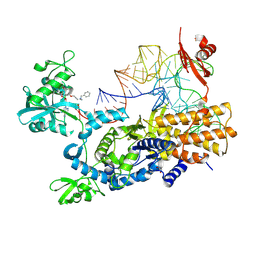 | | Ternary complex of E. coli leucyl-tRNA synthetase, tRNA(leu) and the benzoxaborole cmpd9 in the editing conformation | | Descriptor: | 1,2-ETHANEDIOL, Leucine--tRNA ligase, [(1R,3'S,5S,6R,8R)-3'-(aminomethyl)-8-(6-aminopurin-9-yl)-4'-bromanyl-7'-[3-[methyl-(phenylmethyl)amino]propoxy]spiro[2,4,7-trioxa-3$l^{4}-borabicyclo[3.3.0]octane-3,1'-3H-2,1$l^{4}-benzoxaborole]-6-yl]methyl dihydrogen phosphate, ... | | Authors: | Palencia, A, Hoffmann, G. | | Deposit date: | 2023-07-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Targeting A Microbiota Wolbachian Aminoacyl-tRNA Synthetase To Block Its Pathogenic Host
Science Advances, 2024
|
|
8POS
 
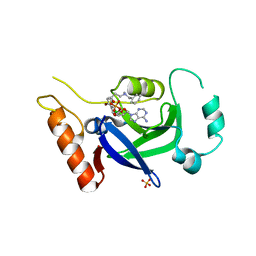 | | Crystal structure of wolbachia leucyl-tRNA synthetase editing domain bound to cmpd9-AMP adduct | | Descriptor: | Leucine--tRNA ligase, SULFATE ION, [(1R,3'S,5S,6R,8R)-3'-(aminomethyl)-8-(6-aminopurin-9-yl)-4'-bromanyl-7'-[3-[methyl-(phenylmethyl)amino]propoxy]spiro[2,4,7-trioxa-3$l^{4}-borabicyclo[3.3.0]octane-3,1'-3H-2,1$l^{4}-benzoxaborole]-6-yl]methyl dihydrogen phosphate | | Authors: | Palencia, A, Hoffmann, G. | | Deposit date: | 2023-07-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | Targeting A Microbiota Wolbachian Aminoacyl-tRNA Synthetase To Block Its Pathogenic Host
Science Advances, 2024
|
|
8POM
 
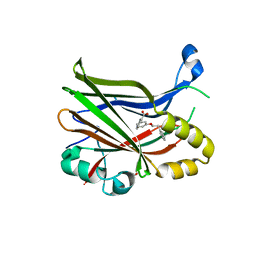 | | TEAD2 in complex with an inhibitor | | Descriptor: | 2-[[3-(2-phenylethoxy)phenyl]amino]pyridine-3-carboxylic acid, GLYCEROL, Transcriptional enhancer factor TEF-4 | | Authors: | Guichou, J.F. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of LM-41 and AF-2112, two flufenamic acid-derived TEAD inhibitors obtained through the replacement of the trifluoromethyl group by aryl rings.
Bioorg.Med.Chem.Lett., 95, 2023
|
|
8POF
 
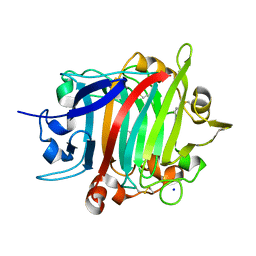 | | The crystal structure of RsSymEG1 reveals a unique form of smaller GH7 endoglucanases alongside GH7 cellobiohydrolases in protist symbionts of termites | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Putative glycosyl hydrolase family7, SODIUM ION | | Authors: | Haataja, T, Sandgren, M, Hansson, H, Stahlberg, J. | | Deposit date: | 2023-07-04 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of RsSymEG1 reveals a unique form of smaller GH7 endoglucanases alongside GH7 cellobiohydrolases in protist symbionts of termites.
Febs J., 291, 2024
|
|
8POE
 
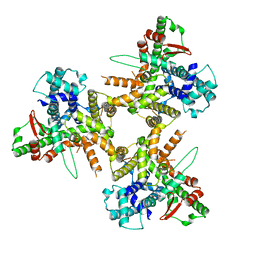 | | Structure of tissue-specific lipid scramblase ATG9B homotrimer, refined with C3 symmetry applied | | Descriptor: | Autophagy-related protein 9B | | Authors: | Chiduza, G.N, Pye, V.E, Tooze, S.A, Cherepanov, P. | | Deposit date: | 2023-07-04 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | ATG9B is a tissue-specific homotrimeric lipid scramblase that can compensate for ATG9A.
Autophagy, 20, 2024
|
|
8POD
 
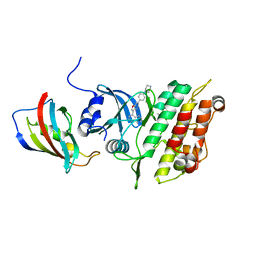 | | Crystal structure of the kinase domain of ACVR1 (ALK2) in complex with FKBP12 and MU1700 | | Descriptor: | 6-(4-piperazin-1-ylphenyl)-3-quinolin-4-yl-furo[3,2-b]pyridine, Activin receptor type-1, FLUORIDE ION, ... | | Authors: | Cros, J, Baltanas-Copado, J, Knapp, S, Paruch, K, Nemec, N, Bullock, A.N. | | Deposit date: | 2023-07-04 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of the kinase domain of ACVR1 (ALK2) in complex with FKBP12 and MU1700
To Be Published
|
|
8POB
 
 | |
8POA
 
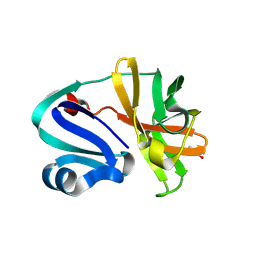 | | Structure of Coxsackievirus A16 (G-10) 2A protease | | Descriptor: | GLYCEROL, Protease 2A, ZINC ION | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-04 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of EV A71 2A protease - to be published
To Be Published
|
|
8PO9
 
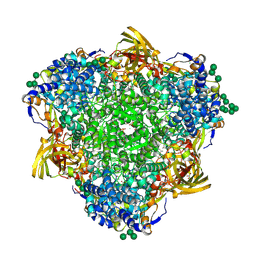 | | Polyethylene oxidation hexamerin PEase Cibeles (XP_026756460) from Galleria mellonella | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Arylphorin, ... | | Authors: | Illanes-Vicioso, R, Ruiz-Lopez, E, Sola, M, Bertocchini, F, Palomo, E.A. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Plastic degradation by insect hexamerins: Near-atomic resolution structures of the polyethylene-degrading proteins from the wax worm saliva.
Sci Adv, 9, 2023
|
|
8PO4
 
 | |
8PO3
 
 | |
8PO2
 
 | |
8PO1
 
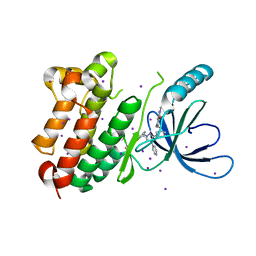 | |
8PO0
 
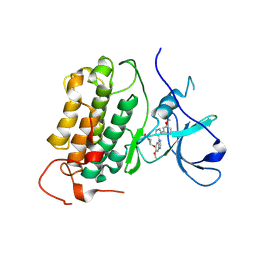 | |
8PNZ
 
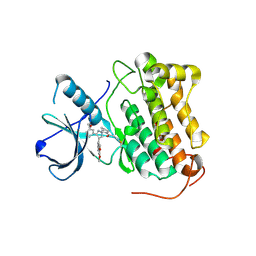 | |
8PNY
 
 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens complexed with phenylhydrazine and in its apo form | | Descriptor: | Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, [6-methyl-5-oxidanyl-4-[(2-phenylhydrazinyl)methyl]pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Expanded Substrate Specificity in D-Amino Acid Transaminases: A Case Study of Transaminase from Blastococcus saxobsidens.
Int J Mol Sci, 24, 2023
|
|
8PNW
 
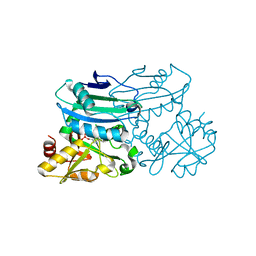 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens in holo form with PLP | | Descriptor: | Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Expanded Substrate Specificity in D-Amino Acid Transaminases: A Case Study of Transaminase from Blastococcus saxobsidens.
Int J Mol Sci, 24, 2023
|
|
8PNO
 
 | |
8PNJ
 
 | | Chorismate mutase | | Descriptor: | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Khatanbaatar, T, Cordara, G, Krengel, U. | | Deposit date: | 2023-06-30 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.355 Å) | | Cite: | Structural analysis of chorismate mutase and cyclohexadienyl dehydratase from Pseudomonas aeruginosa
To Be Published
|
|
8PNE
 
 | | E.coli YihX Wild Type Apo | | Descriptor: | Alpha-D-glucose 1-phosphate phosphatase YihX, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zappala, D, Baumann, P, Jin, Y. | | Deposit date: | 2023-06-30 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Apo structure of E.coli YihX Wild Type
To Be Published
|
|
