2HDV
 
 | |
2HDX
 
 | |
1V9T
 
 | | Structure of E. coli cyclophilin B K163T mutant bound to succinyl-ALA-PRO-ALA-P-nitroanilide | | Descriptor: | (SIN)APA(NIT), cyclophilin B | | Authors: | Konno, M, Sano, Y, Okudaira, K, Kawaguchi, Y, Yamagishi-Ohmori, Y, Fushinobu, S, Matsuzawa, H. | | Deposit date: | 2004-02-03 | | Release date: | 2004-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Escherichia coli cyclophilin B binds a highly distorted form of trans-prolyl peptide isomer
Eur.J.Biochem., 271, 2004
|
|
4NJT
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Yedidi, R.S, Garimella, H, Chang, S.B, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
7G9Z
 
 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z1216822028 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-(propan-2-yl)-1H-pyrrolo[3,2-b]pyridine, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-03 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.622 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1URW
 
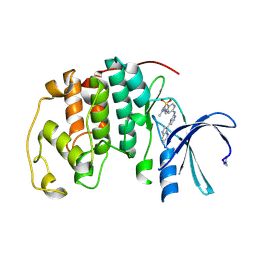 | | CDK2 IN COMPLEX WITH AN IMIDAZO[1,2-b]PYRIDAZINE | | Descriptor: | 2-[4-(N-(3-DIMETHYLAMINOPROPYL)SULPHAMOYL)ANILINO]-, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Byth, K.F, Cooper, N, Culshaw, J.D, Heaton, D.W, Oakes, S.E, Minshull, C.A, Norman, R.A, Pauptit, R.A, Tucker, J.A, Breed, J, Pannifer, A, Rowsell, S, Stanway, J.J, Valentine, A.L, Thomas, A.P. | | Deposit date: | 2003-11-11 | | Release date: | 2004-04-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Imidazo[1,2-B]Pyridazines: A Potent and Selective Class of Cyclin-Dependent Kinase Inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
2XCO
 
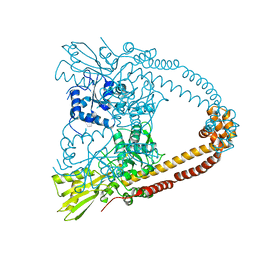 | | The 3.1A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase | | Descriptor: | CALCIUM ION, DNA GYRASE SUBUNIT B, DNA GYRASE SUBUNIT A | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-24 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
2C7W
 
 | | Crystal Structure of human vascular endothelial growth factor-B: Identification of amino acids important for angiogeninc activity | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, VASCULAR ENDOTHELIAL GROWTH FACTOR B PRECURSOR | | Authors: | Iyer, S, Scotney, P.D, Nash, A.D, Acharya, K.R. | | Deposit date: | 2005-11-29 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure of Human Vascular Endothelial Growth Factor-B: Identification of Amino Acids Important for Receptor Binding
J.Mol.Biol., 359, 2006
|
|
4N6H
 
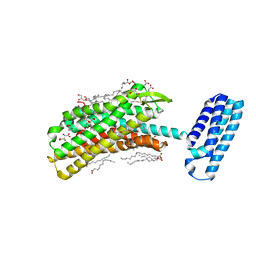 | | 1.8 A Structure of the human delta opioid 7TM receptor (PSI Community Target) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (4bS,8R,8aS,14bR)-7-(cyclopropylmethyl)-5,6,7,8,14,14b-hexahydro-4,8-methano[1]benzofuro[2,3-a]pyrido[4,3-b]carbazole-1,8a(9H)-diol, L(+)-TARTARIC ACID, ... | | Authors: | Fenalti, G, Giguere, P.M, Katritch, V, Huang, X.-P, Thompson, A.A, Han, G.W, Cherezov, V, Roth, B.L, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2013-10-12 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular control of delta-opioid receptor signalling.
Nature, 506, 2014
|
|
1C48
 
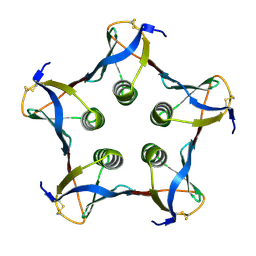 | | MUTATED SHIGA-LIKE TOXIN B SUBUNIT (G62T) | | Descriptor: | PROTEIN (SHIGA-LIKE TOXIN I B SUBUNIT) | | Authors: | Ling, H, Bast, D, Brunton, J.L, Read, R.J. | | Deposit date: | 1999-08-11 | | Release date: | 2000-08-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of the Primary Receptor Binding Site of Shiga-Like Toxin B Subunits: Structures of Mutated Shiga-Like Toxin I B-Pentamer with and without Bound Carbohydrate
To be Published
|
|
4QHM
 
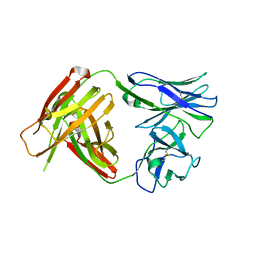 | | I3.1 (unbound) from CH103 Lineage | | Descriptor: | I2 light chain, I3 heavy chain | | Authors: | Fera, D, Harrison, S.C. | | Deposit date: | 2014-05-28 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5UQN
 
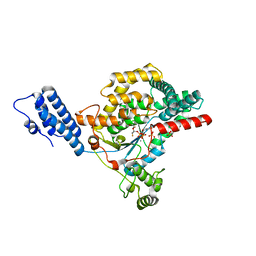 | |
2X7G
 
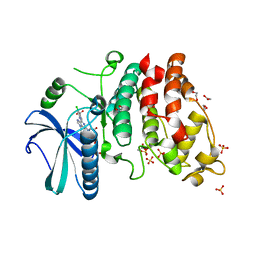 | | Structure of human serine-arginine-rich protein-specific kinase 2 (SRPK2) bound to purvalanol B | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PURVALANOL B, ... | | Authors: | Pike, A.C.W, Savitsky, P, Fedorov, O, Krojer, T, Ugochukwu, E, von Delft, F, Gileadi, O, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Knapp, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Human Serine-Arginine-Rich Protein- Specific Kinase 2 (Srpk2) Bound to Purvalanol B
To be Published
|
|
7H00
 
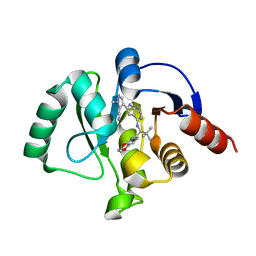 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0011221-001 | | Descriptor: | (4M)-4-(4-{[(1S)-1-(2,3-dihydro[1,4]dioxino[2,3-b]pyridin-6-yl)-2,2-dimethylpropyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-1-methyl-1H-pyrazole-5-carbonitrile, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H0W
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0013738-001 | | Descriptor: | 7-{(1R)-2-methyl-1-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]propyl}-3,4-dihydro-1lambda~6~-thiopyrano[2,3-b]pyridine-1,1(2H)-dione, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.158 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
3B6S
 
 | | Crystal Structure of hla-b*2705 Complexed with the Citrullinated Vasoactive Intestinal Peptide Type 1 Receptor (vipr) Peptide (residues 400-408) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-27 alpha chain, ... | | Authors: | Beltrami, A, Rossmann, M, Fiorillo, M.T, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, A. | | Deposit date: | 2007-10-29 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Citrullination-dependent Differential Presentation of a Self-peptide by HLA-B27 Subtypes.
J.Biol.Chem., 283, 2008
|
|
3BBB
 
 | |
4F36
 
 | |
2VKS
 
 | |
304D
 
 | |
3D29
 
 | | Proteasome Inhibition by Fellutamide B | | Descriptor: | (3R)-3-HYDROXYDODECANOIC ACID, Fellutamide B, PRE10 isoform 1, ... | | Authors: | Groll, M, Hines, J, Fahnestock, M, Crews, M.C. | | Deposit date: | 2008-05-07 | | Release date: | 2008-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Proteasome Inhibition by Fellutamide B Induces Nerve Growth Factor Synthesis
Chem.Biol., 15, 2008
|
|
2ACZ
 
 | | Complex II (Succinate Dehydrogenase) From E. Coli with Atpenin A5 inhibitor co-crystallized at the ubiquinone binding site | | Descriptor: | 3-[(2S,4S,5R)-5,6-DICHLORO-2,4-DIMETHYL-1-OXOHEXYL]-4-HYDROXY-5,6-DIMETHOXY-2(1H)-PYRIDINONE, CARDIOLIPIN, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Horsefield, R, Yankovskaya, V, Sexton, G, Whittingham, W, Shiomi, K, Omura, S, Byrne, B, Cecchini, G, Iwata, S. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction.
J.Biol.Chem., 281, 2006
|
|
2G1A
 
 | | Crystal structure of the complex between Apha class B acid phosphatase/phosphotransferase | | Descriptor: | Class B acid phosphatase, MAGNESIUM ION, {[2-(6-AMINO-9H-PURIN-9-YL)ETHOXY]METHYL}PHOSPHONIC ACID | | Authors: | Leone, R, Calderone, V, Cappelletti, E, Benvenuti, M, Mangani, S. | | Deposit date: | 2006-02-14 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex between Apha class B acid phosphatase/phosphotransferase
To be published
|
|
3QKD
 
 | | Crystal structure of Bcl-xL in complex with a Quinazoline sulfonamide inhibitor | | Descriptor: | (R)-N-(7-(4-((4'-chlorobiphenyl-2-yl)methyl)piperazin-1-yl)quinazolin-4-yl)-4-(4-(dimethylamino)-1-(phenylthio)butan-2-ylamino)-3-nitrobenzenesulfonamide, Bcl-2-like protein 1, CHLORIDE ION, ... | | Authors: | Czabotar, P.E, Smith, B.J. | | Deposit date: | 2011-01-31 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Quinazoline sulfonamides as dual binders of the proteins B-cell lymphoma 2 and B-cell lymphoma extra long with potent proapoptotic cell-based activity.
J.Med.Chem., 54, 2011
|
|
1QUS
 
 | | 1.7 A RESOLUTION STRUCTURE OF THE SOLUBLE LYTIC TRANSGLYCOSYLASE SLT35 FROM ESCHERICHIA COLI | | Descriptor: | 1,2-ETHANEDIOL, BICINE, LYTIC MUREIN TRANSGLYCOSYLASE B, ... | | Authors: | van Asselt, E.J, Dijkstra, A.J, Kalk, K.H, Takacs, B, Keck, W, Dijkstra, B.W. | | Deposit date: | 1999-07-03 | | Release date: | 1999-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Escherichia coli lytic transglycosylase Slt35 reveals a lysozyme-like catalytic domain with an EF-hand.
Structure Fold.Des., 7, 1999
|
|
