5KTS
 
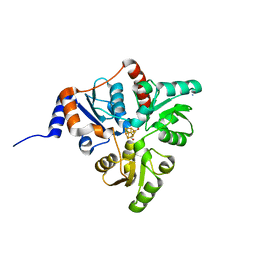 | | Crystal structure of Pyrococcus horikoshii quinolinate synthase (NadA) with bound citraconate and Fe4S4 cluster | | Descriptor: | (~{Z})-2-methylbut-2-enedioic acid, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Ealick, S.E. | | Deposit date: | 2016-07-12 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structures of the Iron-Sulfur Cluster-Dependent Quinolinate Synthase in Complex with Dihydroxyacetone Phosphate, Iminoaspartate Analogues, and Quinolinate.
Biochemistry, 55, 2016
|
|
3E6O
 
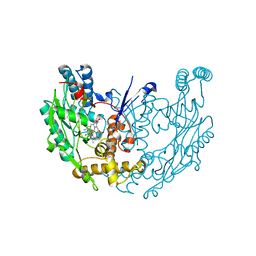 | | Structure of murine INOS oxygenase domain with inhibitor AR-C124355 | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, N-[2-(4-AMINO-5,8-DIFLUORO-1,2-DIHYDROQUINAZOLIN-2-YL)ETHYL]-3-FURAMIDE, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stueh, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-15 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3EMB
 
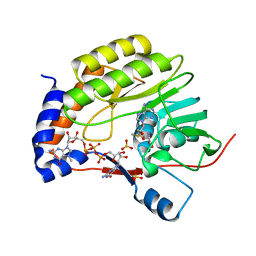 | | Wesselsbron virus Methyltransferase in complex with AdoMet and 7MeGpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Methyltransferase, S-ADENOSYLMETHIONINE, ... | | Authors: | Bollati, M, Ricagno, S, Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2008-09-24 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of RNA Cap in the Wesselsbron Virus NS5 Methyltransferase Domain: Implications for RNA-Capping Mechanisms in Flavivirus
J.Mol.Biol., 385, 2009
|
|
5KUA
 
 | | Cryo-EM reconstruction of Neisseria meningitidis Type IV pilus | | Descriptor: | pilin | | Authors: | Kolappan, S, Coureuil, M, Yu, X, Nassif, X, Craig, L, Egelman, E.H. | | Deposit date: | 2016-07-13 | | Release date: | 2016-10-12 | | Last modified: | 2016-11-30 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of the Neisseria meningitidis Type IV pilus.
Nat Commun, 7, 2016
|
|
5DX5
 
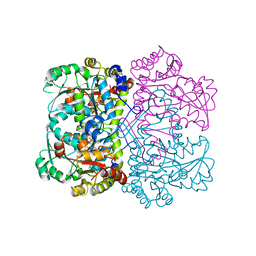 | | Crystal structure of methionine gamma-lyase from Clostridium sporogenes | | Descriptor: | CHLORIDE ION, Methionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Revtovich, S.V, Nikulin, A.D, Morozova, E.A, Anufrieva, N.V, Demidkina, T.V. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure of methionine gamma-lyase from Clostridium sporogenes.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5KWW
 
 | | Crystal Structure of Inhibitor JNJ-53718678 In Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[[5-chloranyl-1-(3-methylsulfonylpropyl)indol-2-yl]methyl]-1-[2,2,2-tris(fluoranyl)ethyl]imidazo[4,5-c]pyridin-2-one, ... | | Authors: | McLellan, J.S, Battles, M.B, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2016-07-19 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Therapeutic efficacy of a respiratory syncytial virus fusion inhibitor.
Nat Commun, 8, 2017
|
|
5DXR
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-{[(3R)-3-methylcyclohexylidene]methanediyl}diphenol | | Descriptor: | 4,4'-{[(3R)-3-methylcyclohexylidene]methanediyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-23 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
1P6M
 
 | | Bovine endothelial NOS heme domain with (4S)-N-(4-amino-5-[aminoethyl]aminopentyl)-N'-nitroguanidine bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
5DYB
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-(3,4-dihydronaphthalen-2(1H)-ylidenemethanediyl)diphenol | | Descriptor: | 4,4'-(3,4-dihydronaphthalen-2(1H)-ylidenemethanediyl)diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-24 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
2ITF
 
 | | Crystal structure IsdA NEAT domain from Staphylococcus aureus with heme bound | | Descriptor: | Iron-regulated surface determinant protein A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Grigg, J.C, Vermeiren, C.L, Heinrichs, D.E, Murphy, M.E. | | Deposit date: | 2006-10-19 | | Release date: | 2006-12-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Haem recognition by a Staphylococcus aureus NEAT domain.
Mol.Microbiol., 63, 2007
|
|
1RZZ
 
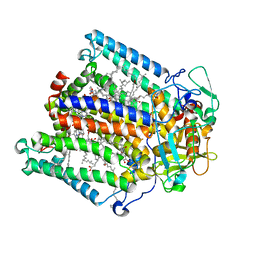 | | PHOTOSYNTHETIC REACTION CENTER DOUBLE MUTANT FROM RHODOBACTER SPHAEROIDES WITH ASP L213 REPLACED WITH ASN AND ARG M233 REPLACED WITH CYS IN THE CHARGE-NEUTRAL DQAQB STATE (TETRAGONAL FORM) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Xu, Q, Axelrod, H.L, Abresch, E.C, Paddock, M.L, Okamura, M.Y, Feher, G. | | Deposit date: | 2003-12-29 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-Ray Structure Determination of Three Mutants of the Bacterial Photosynthetic Reaction Centers from Rb. sphaeroides; Altered Proton Transfer Pathways.
STRUCTURE, 12, 2004
|
|
1EG9
 
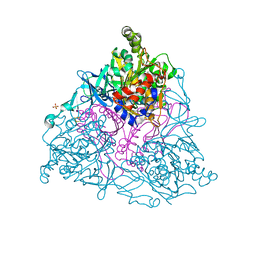 | | NAPHTHALENE 1,2-DIOXYGENASE WITH INDOLE BOUND IN THE ACTIVE SITE. | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, INDOLE, ... | | Authors: | Carredano, E, Karlsson, A, Kauppi, B, Choudhury, D, Parales, R.E, Parales, J.V, Lee, K, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2000-02-15 | | Release date: | 2000-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate binding site of naphthalene 1,2-dioxygenase: functional implications of indole binding.
J.Mol.Biol., 296, 2000
|
|
2DM9
 
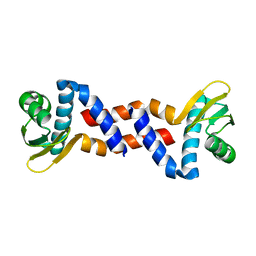 | |
4CYI
 
 | | Chaetomium thermophilum Pan3 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN3-LIKE PROTEIN, ... | | Authors: | Wolf, J, Valkov, E, Allen, M.D, Meineke, B, Gordiyenko, Y, McLaughlin, S.H, Olsen, T.M, Robinson, C.V, Bycroft, M, Stewart, M, Passmore, L.A. | | Deposit date: | 2014-04-11 | | Release date: | 2014-06-11 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis for Pan3 Binding to Pan2 and its Function in Mrna Recruitment and Deadenylation
Embo J., 33, 2014
|
|
1OTU
 
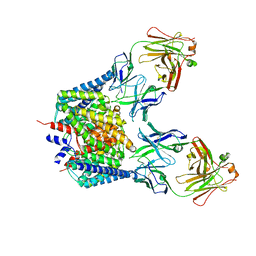 | | Structure of the Escherichia coli ClC Chloride channel E148Q mutant and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (Heavy chain), Fab fragment (Light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-23 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
7KD3
 
 | |
2ZBJ
 
 | | Crystal structure of Dioclea rostrata lectin | | Descriptor: | CALCIUM ION, Lectin alpha chain, MANGANESE (II) ION, ... | | Authors: | de Oliveira, T.M, Delatorre, P, da Rocha, B.A.M, de Sousa, E.P, Nascimento, K.S, Bezerra, G.A, Moura, T.R, Benevides, R.G, Bezerra, E.H.S, Moreno, F.B.M.B, Freire, V.N, de Azevedo Jr, W.F, Cavada, B.S. | | Deposit date: | 2007-10-22 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Dioclea rostrata lectin: Insights into understanding the pH-dependent dimer-tetramer equilibrium and the structural basis for carbohydrate recognition in Diocleinae lectins
J.Struct.Biol., 164, 2008
|
|
5KO8
 
 | | Crystal structure of haliscomenobacter hydrossis iodotyrosine deiodinase (IYD) bound to FMN and mono-iodotyrosine (I-Tyr) | | Descriptor: | 3-IODO-TYROSINE, FLAVIN MONONUCLEOTIDE, Nitroreductase | | Authors: | Ingavat, N, Kavran, J.M, Sun, Z, Rokita, S.E. | | Deposit date: | 2016-06-29 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Active Site Binding Is Not Sufficient for Reductive Deiodination by Iodotyrosine Deiodinase.
Biochemistry, 56, 2017
|
|
3HNK
 
 | | Crystal structure of the dimeric assembly of the cyt cb562 variant RIDC-1 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562 | | Authors: | Salgado, E.N, Lewis, R.A, Brodin, J, Tezcan, F.A. | | Deposit date: | 2009-05-31 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Metal templated design of protein interfaces.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5XXJ
 
 | |
4HST
 
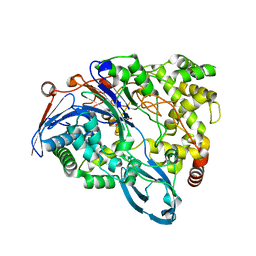 | | Crystal structure of a double mutant of a class III engineered cephalosporin acylase | | Descriptor: | 5,5-dihydroxy-L-norvaline, glutaryl-7-aminocephalosporanic acid acylase alpha chain, glutaryl-7-aminocephalosporanic acid acylase beta chain | | Authors: | Vrielink, A, Golden, E, Patterson, R, Tie, W.J, Anandan, A, Flematti, G, Molla, G, Rosini, E, Pollegioni, L. | | Deposit date: | 2012-10-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Structure of a class III engineered cephalosporin acylase: comparisons with class I acylase and implications for differences in substrate specificity and catalytic activity.
Biochem.J., 451, 2013
|
|
5DOS
 
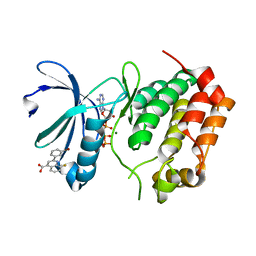 | | Aurora A Kinase in Complex with AA35 and ATP in Space Group P6122 | | Descriptor: | 2-(3-bromophenyl)-8-fluoroquinoline-4-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, ... | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-11 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
5EDK
 
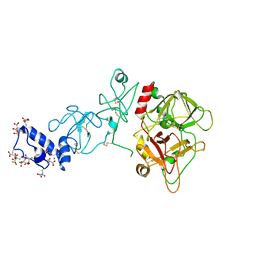 | | Crystal structure of prothrombin deletion mutant residues 146-167 ( Form II ). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Pozzi, N, Chen, Z, Di Cera, E. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.214 Å) | | Cite: | How the Linker Connecting the Two Kringles Influences Activation and Conformational Plasticity of Prothrombin.
J.Biol.Chem., 291, 2016
|
|
4CVN
 
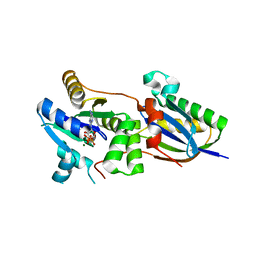 | | Structure of the Fap7-Rps14 complex | | Descriptor: | 30S RIBOSOMAL PROTEIN S11, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Loch, J, Blaud, M, Rety, S, Lebaron, S, Deschamps, P, Bareille, J, Jombart, J, Robert-Paganin, J, Delbos, L, Chardon, F, Zhang, E, Charenton, C, Tollervey, D, Leulliot, N. | | Deposit date: | 2014-03-28 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | RNA Mimicry by the Fap7 Adenylate Kinase in Ribosome Biogenesis
Plos Biol., 12, 2014
|
|
5L7V
 
 | | Crystal Structure of BvGH123 with bond transition state analog Galthiazoline. | | Descriptor: | (3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, glycoside hydrolase | | Authors: | Roth, C, Petricevic, M, John, A, Goddard-Borger, E.D, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mechanistic insights into a Bacteroides vulgatus retaining N-acetyl-beta-galactosaminidase that uses neighbouring group participation.
Chem. Commun. (Camb.), 52, 2016
|
|
