1RPC
 
 | | SOLUTION STRUCTURE OF RP 71955, A NEW 21 AMINO ACID TRICYCLIC PEPTIDE ACTIVE AGAINST HIV-1 VIRUS | | Descriptor: | Tricyclic peptide RP 71955 | | Authors: | Frechet, D, Guitton, J.D, Herman, F, Faucher, D, Helynck, G, Monegier Du Sorbier, B, Ridoux, J.P, James-Surcouf, E, Vuilhorgne, M. | | Deposit date: | 1993-08-31 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RP 71955, a new 21 amino acid tricyclic peptide active against HIV-1 virus.
Biochemistry, 33, 1994
|
|
1RPE
 
 | | THE PHAGE 434 OR2/R1-69 COMPLEX AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*AP*CP*AP*AP*GP*AP*TP*AP*CP*AP*TP*TP*G P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*CP*AP*AP*TP*GP*TP*AP*TP*CP*TP*TP*GP*T P*TP*TP*G)-3'), PROTEIN (434 REPRESSOR) | | Authors: | Shimon, L.J.W, Harrison, S.C. | | Deposit date: | 1993-03-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The phage 434 OR2/R1-69 complex at 2.5 A resolution.
J.Mol.Biol., 232, 1993
|
|
1RPF
 
 | |
1RPG
 
 | | STRUCTURES OF RNASE A COMPLEXED WITH 3'-CMP AND D(CPA): ACTIVE SITE CONFORMATION AND CONSERVED WATER MOLECULES | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2'-DEOXYCYTIDINE-2'-DEOXYADENOSINE-3',5'-MONOPHOSPHATE, RIBONUCLEASE A | | Authors: | Zegers, I, Wyns, L, Palmer, R. | | Deposit date: | 1994-08-29 | | Release date: | 1994-12-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structures of RNase A complexed with 3'-CMP and d(CpA): active site conformation and conserved water molecules.
Protein Sci., 3, 1994
|
|
1RPH
 
 | |
1RPI
 
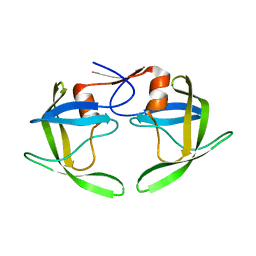 | | Crystal structures of a Multidrug-Resistant HIV-1 Protease Reveal an Expanded Active Site Cavity | | Descriptor: | alpha-D-glucopyranose, protease | | Authors: | Logsdon, B.C, Vickrey, J.F, Martin, P, Proteasa, G, Koepke, J.I, Terlecky, S.R, Wawrzak, Z, Winters, M.A, Merigan, T.C, Kovari, L.C. | | Deposit date: | 2003-12-03 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of a multidrug-resistant human immunodeficiency virus type 1 protease reveal an expanded active-site cavity.
J.Virol., 78, 2004
|
|
1RPJ
 
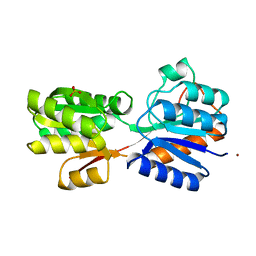 | | CRYSTAL STRUCTURE OF D-ALLOSE BINDING PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | PROTEIN (PRECURSOR OF PERIPLASMIC SUGAR RECEPTOR), SULFATE ION, ZINC ION, ... | | Authors: | Chaudhuri, B, Jones, T.A, Mowbray, S.L. | | Deposit date: | 1999-02-04 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of D-allose binding protein from Escherichia coli bound to D-allose at 1.8 A resolution.
J.Mol.Biol., 286, 1999
|
|
1RPK
 
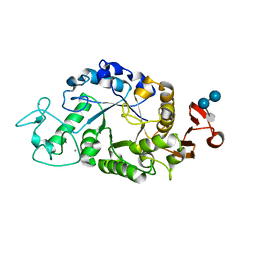 | | Crystal structure of barley alpha-amylase isozyme 1 (amy1) in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-amylase type 1 isozyme, ... | | Authors: | Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2003-12-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide Binding to Barley {alpha}-Amylase 1
J.Biol.Chem., 280, 2005
|
|
1RPL
 
 | |
1RPM
 
 | |
1RPN
 
 | | Crystal Structure of GDP-D-mannose 4,6-dehydratase in complexes with GDP and NADPH | | Descriptor: | GDP-mannose 4,6-dehydratase, GUANOSINE-5'-DIPHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Webb, N.A, Mulichak, A.M, Lam, J.S, Rocchetta, H.L, Garavito, R.M. | | Deposit date: | 2003-12-03 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of a tetrameric GDP-D-mannose 4,6-dehydratase from a bacterial GDP-D-rhamnose biosynthetic pathway.
Protein Sci., 13, 2004
|
|
1RPO
 
 | |
1RPQ
 
 | | High Affinity IgE Receptor (alpha chain) Complexed with Tight-Binding E131 'zeta' Peptide from Phage Display | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stamos, J, Eigenbrot, C, Nakamura, G.R, Reynolds, M.E, Yin, J.P, Lowman, H.B, Fairbrother, W.J, Starovasnik, M.A. | | Deposit date: | 2003-12-03 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Convergent Recognition of the IgE Binding Site on the High-Affinity IgE Receptor.
Structure, 12, 2004
|
|
1RPR
 
 | | THE STRUCTURE OF COLE1 ROP IN SOLUTION | | Descriptor: | ROP | | Authors: | Eberle, W, Pastore, A, Klaus, W, Sander, C, Roesch, P. | | Deposit date: | 1991-10-09 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of ColE1 rop in solution.
J.Biomol.NMR, 1, 1991
|
|
1RPS
 
 | | Crystallographic Analysis of the Interaction of Nitric Oxide with Quaternary-T Human Hemoglobin. Hemoglobin exposed to NO under anerobic conditions | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, NITRIC OXIDE, ... | | Authors: | Chan, N.-L, Kavanaugh, J.S, Rogers, P.H, Arnone, A. | | Deposit date: | 2003-12-03 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystallographic analysis of the interaction of nitric oxide with quaternary-T human hemoglobin.
Biochemistry, 43, 2004
|
|
1RPT
 
 | |
1RPU
 
 | | Crystal Structure of CIRV p19 bound to siRNA | | Descriptor: | 19 kDa protein, 5'-R(P*CP*GP*UP*AP*CP*GP*CP*GP*UP*CP*AP*CP*GP*CP*GP*UP*AP*CP*GP*UP*U)-3' | | Authors: | Vargason, J.M, Szittya, G, Burgyan, J, Hall, T.M.T. | | Deposit date: | 2003-12-03 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Size selective recognition of siRNA by an RNA silencing suppressor
Cell(Cambridge,Mass.), 115, 2003
|
|
1RPV
 
 | | HIV-1 REV PROTEIN (RESIDUES 34-50) | | Descriptor: | HIV-1 REV PROTEIN | | Authors: | Scanlon, M.J, Fairlie, D.P, Craik, D.J, Englebretsen, D.R, West, M.L. | | Deposit date: | 1995-05-04 | | Release date: | 1995-10-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the RNA-binding peptide from human immunodeficiency virus (type 1) Rev.
Biochemistry, 34, 1995
|
|
1RPW
 
 | | Crystal Structure Of The Multidrug Binding Protein Qacr Bound To The Diamidine Hexamidine | | Descriptor: | 4,4'[1,6-HEXANEDIYLBIS(OXY)]BISBENZENECARBOXIMIDAMIDE, SULFATE ION, Transcriptional regulator qacR | | Authors: | Murray, D.S, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2003-12-03 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of QacR-diamidine complexes reveal additional multidrug-binding modes and a novel mechanism of drug charge neutralization.
J.Biol.Chem., 279, 2004
|
|
1RPX
 
 | |
1RPY
 
 | | CRYSTAL STRUCTURE OF THE DIMERIC SH2 DOMAIN OF APS | | Descriptor: | SULFATE ION, adaptor protein APS | | Authors: | Hu, J, Liu, J, Ghirlando, R, Saltiel, A.R, Hubbard, S.R. | | Deposit date: | 2003-12-03 | | Release date: | 2003-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recruitment of the adaptor protein APS to the activated insulin receptor.
Mol.Cell, 12, 2003
|
|
1RPZ
 
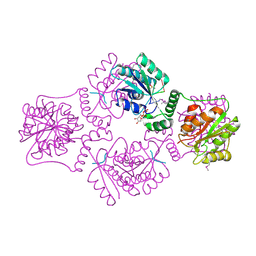 | | T4 POLYNUCLEOTIDE KINASE BOUND TO 5'-TGCAC-3' SSDNA | | Descriptor: | 5'-D(*TP*GP*CP*AP*C)-3', ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Eastberg, J.H, Pelletier, J, Stoddard, B.L. | | Deposit date: | 2003-12-03 | | Release date: | 2004-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition of DNA substrates by T4 bacteriophage polynucleotide kinase.
Nucleic Acids Res., 32, 2004
|
|
1RQ0
 
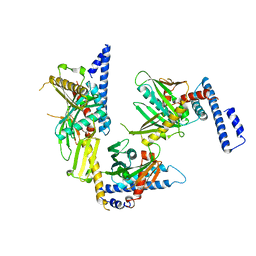 | | Crystal structure of peptide releasing factor 1 | | Descriptor: | Peptide chain release factor 1 | | Authors: | Shin, D.H, Brandsen, J, Jancarik, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-12-03 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural analyses of peptide release factor 1 from Thermotoga maritima reveal domain flexibility required for its interaction with the ribosome.
J.Mol.Biol., 341, 2004
|
|
1RQ1
 
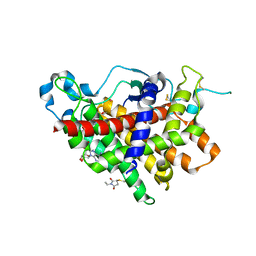 | | Structure of Ero1p, Source of Disulfide Bonds for Oxidative Protein Folding in the Cell | | Descriptor: | 1-ETHYL-PYRROLIDINE-2,5-DIONE, CADMIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Gross, E, Kastner, D.B, Kaiser, C.A, Fass, D. | | Deposit date: | 2003-12-04 | | Release date: | 2004-06-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of ero1p, source of disulfide bonds for oxidative protein folding in the cell.
Cell(Cambridge,Mass.), 117, 2004
|
|
1RQ2
 
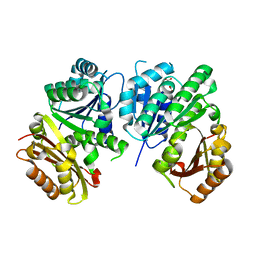 | | MYCOBACTERIUM TUBERCULOSIS FTSZ IN COMPLEX WITH CITRATE | | Descriptor: | CITRIC ACID, Cell division protein ftsZ | | Authors: | Leung, A.K.W, White, E.L, Ross, L.J, Reynolds, R.C, DeVito, J.A, Borhani, D.W. | | Deposit date: | 2003-12-04 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches.
J.Mol.Biol., 342, 2004
|
|
