2KMK
 
 | | Gfi-1 Zinc Fingers 3-5 complexed with DNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*AP*AP*AP*TP*CP*AP*CP*TP*GP*CP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*GP*CP*AP*GP*TP*GP*AP*TP*TP*TP*AP*TP*G)-3'), ZINC ION, ... | | Authors: | Lee, S, Wu, Z. | | Deposit date: | 2009-07-30 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Gfi-1 zinc domain bound to consensus DNA.
J.Mol.Biol., 397, 2010
|
|
7KIK
 
 | | Wheat dwarf virus Rep domain circular permutation complexed with a single-stranded DNA 10-mer comprising the cleavage site and Mn2+ | | Descriptor: | DNA (5'-D(*TP*AP*AP*TP*AP*TP*TP*AP*CP*C)-3'), MANGANESE (II) ION, Replication-associated protein | | Authors: | Tompkins, K.J, Gordon, W.R, Shi, K, Litzau, L.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Watson-Crick Base-Pairing Requirements for ssDNA Recognition and Processing in Replication-Initiating HUH Endonucleases.
Mbio, 14, 2023
|
|
7OBN
 
 | | Structural investigations of a new L3 DNA ligase: structure-function analysis | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3'), DNA ligase, ... | | Authors: | Leiros, H.-K.S, Williamson, A. | | Deposit date: | 2021-04-23 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Bacteriophage origin of some minimal ATP-dependent DNA ligases: a new structure from Burkholderia pseudomallei with striking similarity to Chlorella virus ligase.
Sci Rep, 11, 2021
|
|
6TUX
 
 | | human XPG-DNA, Complex 2 | | Descriptor: | DNA (5'-D(P*AP*AP*CP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*GP*AP*GP*TP*T)-3'), DNA repair protein complementing XP-G cells,DNA repair protein complementing XP-G cells | | Authors: | Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2020-01-08 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of human XPG, the xeroderma pigmentosum group G endonuclease, provides insight into nucleotide excision DNA repair.
Nucleic Acids Res., 48, 2020
|
|
4HIG
 
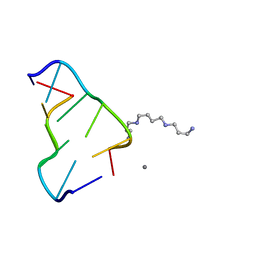 | | Ultrahigh-resolution crystal structure of Z-DNA in complex with Mn2+ ion. | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MANGANESE (II) ION, SPERMINE (FULLY PROTONATED FORM) | | Authors: | Drozdzal, P, Gilski, M, Kierzek, R, Lomozik, L, Jaskolski, M. | | Deposit date: | 2012-10-11 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | Ultrahigh-resolution crystal structures of Z-DNA in complex with Mn(2+) and Zn(2+) ions.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6TUW
 
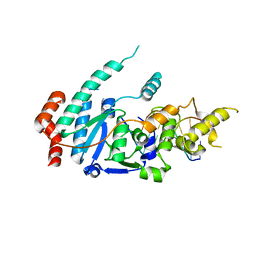 | | human XPG-DNA, Complex 1 | | Descriptor: | DNA (5'-D(P*GP*AP*AP*CP*TP*CP*TP*G)-3'), DNA (5'-D(P*TP*GP*CP*AP*GP*AP*GP*TP*TP*C)-3'), DNA repair protein complementing XP-G cells,DNA repair protein complementing XP-G cells | | Authors: | Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2020-01-08 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The crystal structure of human XPG, the xeroderma pigmentosum group G endonuclease, provides insight into nucleotide excision DNA repair.
Nucleic Acids Res., 48, 2020
|
|
3BNA
 
 | |
5GKP
 
 | |
7NS0
 
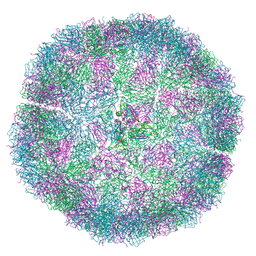 | | Bacilladnavirus capsid structure | | Descriptor: | Capsid protein VP2 | | Authors: | Munke, A, Okamoto, K. | | Deposit date: | 2021-03-05 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Primordial Capsid and Spooled ssDNA Genome Structures Unravel Ancestral Events of Eukaryotic Viruses.
Mbio, 13, 2022
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
6P7B
 
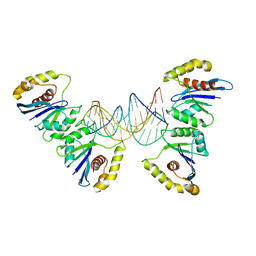 | | Crystal structure of Fowlpox virus resolvase and substrate Holliday junction DNA complex | | Descriptor: | DNA (29-MER), Holliday junction resolvase | | Authors: | Li, N, Shi, K, Rao, T, Banerjee, S, Aihara, H. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.317 Å) | | Cite: | Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase.
Sci Rep, 10, 2020
|
|
8SSQ
 
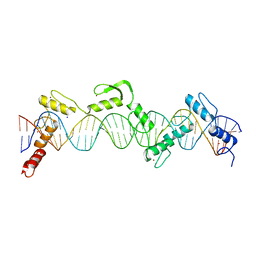 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 35mer DNA 35-4 | | Descriptor: | DNA (35-MER) Strand 2, DNA (35-MER) Strand I, SODIUM ION, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSR
 
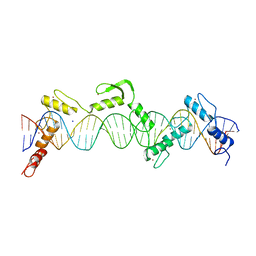 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 35mer DNA 35-20 | | Descriptor: | DNA (35-MER) Strand I, DNA (35-MER) Strand II, SODIUM ION, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
6J44
 
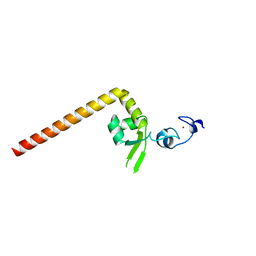 | | Crystal structure of the redefined DNA-binding domain of human XPA | | Descriptor: | DNA repair protein complementing XP-A cells, ZINC ION | | Authors: | Lian, F.M, Yang, X, Yang, W, Jiang, Y.L, Qian, C. | | Deposit date: | 2019-01-07 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural characterization of the redefined DNA-binding domain of human XPA.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
8SVD
 
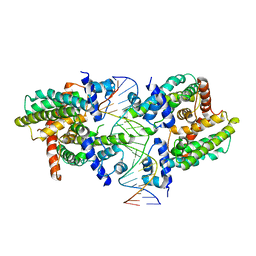 | |
4IQJ
 
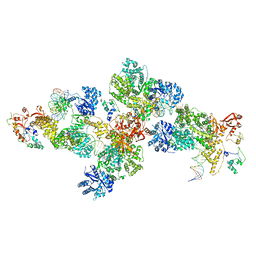 | | Structure of PolIIIalpha-Tauc-DNA complex suggests an atomic model of the replisome | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*GP*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*CP*G)-3'), DNA (5'-D(P*CP*GP*AP*AP*AP*CP*GP*AP*CP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*A)-3'), DNA (5'-D(P*CP*GP*AP*AP*AP*CP*GP*AP*CP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*AP*(DOC))-3'), ... | | Authors: | Liu, B, Lin, J, Steitz, T. | | Deposit date: | 2013-01-11 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of PolIIIalpha-Tauc-DNA complex suggests an atomic model of the replisome
Structure, 21, 2013
|
|
6GCD
 
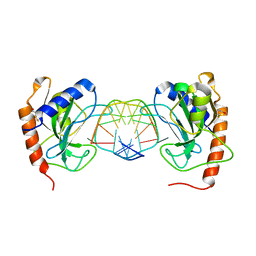 | |
2VYE
 
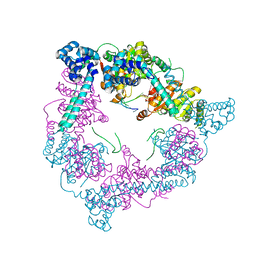 | | Crystal Structure of the DnaC-ssDNA complex | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP)-3', REPLICATIVE DNA HELICASE | | Authors: | Lo, Y.H, Tsai, K.L, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2008-07-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | The Crystal Structure of a Replicative Hexameric Helicase Dnac and its Complex with Single-Stranded DNA.
Nucleic Acids Res., 37, 2009
|
|
6HC3
 
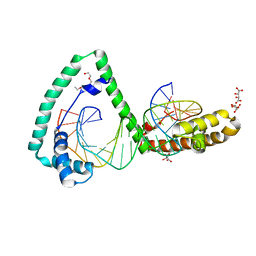 | | TFAM bound to Site-X | | Descriptor: | DNA (5'-D(*TP*TP*TP*GP*GP*TP*GP*GP*AP*AP*AP*TP*TP*TP*TP*TP*TP*GP*TP*TP*AP*G)-3'), DNA/RNA (5'-D(*TP*AP*AP*CP*AP*AP*AP*AP*AP*AP*TP*TP*TP*CP*CP*AP*CP*CP*AP*AP*AP*C)-3'), L(+)-TARTARIC ACID, ... | | Authors: | Fernandez-Millan, P, Cuppari, A, Tarres-Sole, A, Rubio-Cosials, A, Lyonnais, S, Sola, M. | | Deposit date: | 2018-08-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | DNA specificities modulate the binding of human transcription factor A to mitochondrial DNA control region.
Nucleic Acids Res., 47, 2019
|
|
8HTX
 
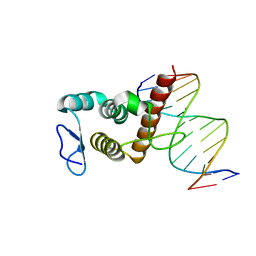 | | Crystal structure of BANP in complex with methylated DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*(5CM)P*GP*CP*GP*AP*GP*AP*G)-3'), Protein BANP | | Authors: | Zhang, J, Min, J, Liu, K. | | Deposit date: | 2022-12-21 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
6D9L
 
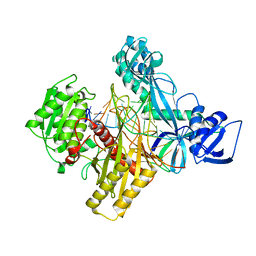 | | Ternary RsAgo Complex with Guide RNA and Target DNA Containing G-A Non-canonical Pair | | Descriptor: | DNA (5'-D(P*TP*CP*GP*TP*CP*AP*CP*CP*TP*GP*AP*GP*CP*AP*GP*TP*AP*AP*C)-3'), MAGNESIUM ION, RNA (5'-R(P*UP*UP*AP*CP*UP*GP*CP*GP*CP*AP*GP*GP*UP*GP*AP*CP*GP*A)-3'), ... | | Authors: | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | Deposit date: | 2018-04-30 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
8AK4
 
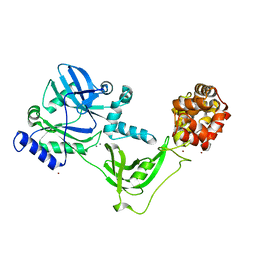 | | Structure of the C-terminally truncated NAD+-dependent DNA ligase from the poly-extremophile Deinococcus radiodurans | | Descriptor: | DNA ligase, MANGANESE (II) ION, ZINC ION | | Authors: | Fernandes, A, Williamson, A.K, Matias, P.M, Moe, E. | | Deposit date: | 2022-07-29 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structure/function studies of the NAD + -dependent DNA ligase from the poly-extremophile Deinococcus radiodurans reveal importance of the BRCT domain for DNA binding.
Extremophiles, 27, 2023
|
|
1LQM
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Varshney, U, Vijayan, M. | | Deposit date: | 2002-05-10 | | Release date: | 2002-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3MDC
 
 | | DNA polymerase lambda in complex with dFdCTP | | Descriptor: | 2'-deoxy-2',2'-difluorocytidine 5'-(tetrahydrogen triphosphate), DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*CP*GP*GP*CP*GP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Garcia-Diaz, M, Murray, M, Kunkel, T, Chou, K.M. | | Deposit date: | 2010-03-30 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Interaction between DNA Polymerase lambda and anticancer nucleoside analogs.
J.Biol.Chem., 285, 2010
|
|
1LQG
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Handa, P, Varshney, U, Vijayan, M. | | Deposit date: | 2002-05-10 | | Release date: | 2002-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
