4M3V
 
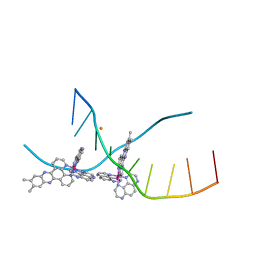 | | X-ray crystal structure of the ruthenium complex [Ru(Tap)2(dppz-{Me2})]2+ bound to d(TCGGTACCGA) | | Descriptor: | (11,12-dimethyldipyrido[3,2-a:2',3'-c]phenazine-kappa~2~N~4~,N~5~)[bis(pyrazino[2,3-f]quinoxaline-kappa~2~N~1~,N~10~)]ruthenium(2+), BARIUM ION, DNA decamer sequence | | Authors: | Niyazi, H, Teixeira, S, Mitchell, E, Forsyth, T, Cardin, C. | | Deposit date: | 2013-08-06 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray crystal structure of the ruthenium complex [Ru(Tap)2(dppz-{Me2})]2+ bound to d(TCGGTACCGA)
To be Published
|
|
7N8K
 
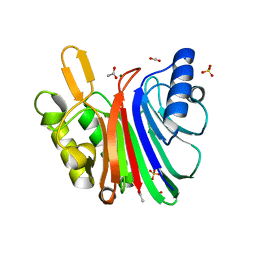 | | LINE-1 endonuclease domain complex with Mg | | Descriptor: | ACETATE ION, LINE-1 retrotransposable element ORF2 protein, MAGNESIUM ION, ... | | Authors: | Korolev, S, Miller, I. | | Deposit date: | 2021-06-15 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural dissection of sequence recognition and catalytic mechanism of human LINE-1 endonuclease.
Nucleic Acids Res., 49, 2021
|
|
7MSV
 
 | |
8TWH
 
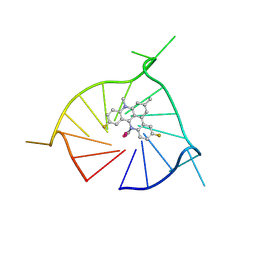 | | Crystal structure of (GGGTT)3GGG G-quadruplex in complex with small molecule ligand RHPS4 | | Descriptor: | (GGGTT)3GGG DNA, 3,11-DIFLUORO-6,8,13-TRIMETHYL-8H-QUINO[4,3,2-KL]ACRIDIN-13-IUM, POTASSIUM ION | | Authors: | Yatsunyk, L.A, Martin, K.N, Lam, G. | | Deposit date: | 2023-08-21 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of (GGGTT)3GGG G-quadruplex in complex with small molecule ligand RHPS4
To be published
|
|
3K2P
 
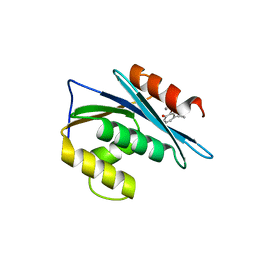 | | HIV-1 Reverse Transcriptase Isolated RnaseH Domain with the Inhibitor beta-thujaplicinol Bound at the Active Site | | Descriptor: | 2,7-dihydroxy-4-(propan-2-yl)cyclohepta-2,4,6-trien-1-one, MANGANESE (II) ION, Reverse Transcriptase | | Authors: | Pauly, T.A, Himmel, D.M, Maegley, K, Arnold, E. | | Deposit date: | 2009-09-30 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of HIV-1 reverse transcriptase with the inhibitor beta-Thujaplicinol bound at the RNase H active site.
Structure, 17, 2009
|
|
4GY5
 
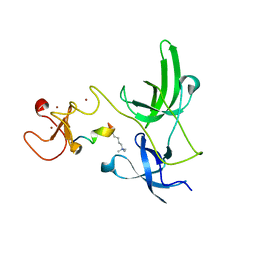 | | Crystal structure of the tandem tudor domain and plant homeodomain of UHRF1 with Histone H3K9me3 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Peptide from Histone H3.3, ZINC ION | | Authors: | Cheng, J, Yang, Y, Fang, J, Xiao, J, Zhu, T, Chen, F, Wang, P, Xu, Y. | | Deposit date: | 2012-09-05 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.956 Å) | | Cite: | Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein
J.Biol.Chem., 288, 2013
|
|
1Y6U
 
 | |
6FY5
 
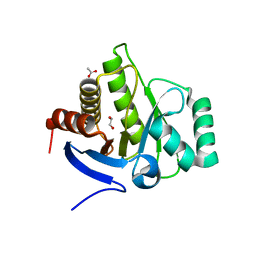 | | Crystal structure of the macro domain of human macroh2a2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Core histone macro-H2A.2 | | Authors: | Hothorn, M. | | Deposit date: | 2018-03-10 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | MacroH2A histone variants limit chromatin plasticity through two distinct mechanisms.
EMBO Rep., 19, 2018
|
|
8FOM
 
 | | Crystal structure of tRNA^Lys(SUU) bound to UAA codon in the ribosomal P site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.A, Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2023-01-02 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Structural basis for reduced ribosomal A-site fidelity in response to P-site codon-anticodon mismatches.
J.Biol.Chem., 299, 2023
|
|
3ZR1
 
 | | Crystal structure of human MTH1 | | Descriptor: | 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, ACETATE ION, SULFATE ION | | Authors: | Svensson, L.M, Jemth, A, Desroses, M, Loseva, O, Helleday, T, Hogbom, M, Stenmark, P. | | Deposit date: | 2011-06-13 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Human Mth1 and the 8-Oxo-Dgmp Product Complex.
FEBS Lett., 585, 2011
|
|
3I2S
 
 | | Crystal structure of the hairpin ribozyme with a 2'OMe substrate and N1-deazaadenosine at position A10 | | Descriptor: | 5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*GP*U)-3', 5'-R(*UP*CP*GP*UP*GP*GP*UP*AP*CP*AP*UP*UP*AP*CP*CP*UP*GP*CP*C)-3', COBALT HEXAMMINE(III), ... | | Authors: | Wedekind, J.E, Spitale, R.C, Krucinska, J. | | Deposit date: | 2009-06-29 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Single-atom imino substitutions at A9 and A10 reveal distinct effects on the fold and function of the hairpin ribozyme catalytic core.
Biochemistry, 48, 2009
|
|
6IE8
 
 | | RamR in complex with cholic acid | | Descriptor: | CHOLIC ACID, Regulatory protein, SULFATE ION | | Authors: | Nakashima, R, Sakurai, K, Yamasaki, S, Nishino, K. | | Deposit date: | 2018-09-13 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the multidrug resistance regulator RamR complexed with bile acids.
Sci Rep, 9, 2019
|
|
4NR3
 
 | |
3I2R
 
 | | Crystal structure of the hairpin ribozyme with a 2',5'-linked substrate with N1-deazaadenosine at position A9 | | Descriptor: | 5'-R(*UP*CP*CP*CP*AP*GP*UP*CP*CP*AP*CP*CP*GP*U)-3', 5'-R(*UP*CP*GP*UP*GP*GP*UP*AP*CP*AP*UP*UP*AP*CP*CP*UP*GP*CP*C)-3', COBALT HEXAMMINE(III), ... | | Authors: | Wedekind, J.E, Spitale, R.C, Krucinska, J. | | Deposit date: | 2009-06-29 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Single-atom imino substitutions at A9 and A10 reveal distinct effects on the fold and function of the hairpin ribozyme catalytic core.
Biochemistry, 48, 2009
|
|
3I2Q
 
 | | Crystal structure of the hairpin ribozyme with 2'OMe substrate strand and N1-deazaadenosine at position A9 | | Descriptor: | 5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*GP*U)-3', 5'-R(*UP*CP*GP*UP*GP*GP*UP*AP*CP*AP*UP*UP*AP*CP*CP*UP*GP*CP*C)-3', COBALT HEXAMMINE(III), ... | | Authors: | Wedekind, J.E, Spitale, R.C, Krucinska, J. | | Deposit date: | 2009-06-29 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Single-atom imino substitutions at A9 and A10 reveal distinct effects on the fold and function of the hairpin ribozyme catalytic core.
Biochemistry, 48, 2009
|
|
3I2U
 
 | | Crystal structure of the haiprin ribozyme with a 2',5'-linked substrate and N1-deazaadenosine at position A10 | | Descriptor: | 5'-R(*UP*CP*CP*CP*AP*GP*UP*CP*CP*AP*CP*CP*GP*U)-3', 5'-R(*UP*CP*GP*UP*GP*GP*UP*AP*CP*AP*UP*UP*AP*CP*CP*UP*GP*CP*C)-3', COBALT HEXAMMINE(III), ... | | Authors: | Wedekind, J.E, Spitale, R.C, Krucinska, J. | | Deposit date: | 2009-06-29 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Single-atom imino substitutions at A9 and A10 reveal distinct effects on the fold and function of the hairpin ribozyme catalytic core.
Biochemistry, 48, 2009
|
|
3IFM
 
 | |
8U3S
 
 | |
1THC
 
 | | CRYSTAL STRUCTURE DETERMINATION AT 2.3A OF HUMAN TRANSTHYRETIN-3',5'-DIBROMO-2',4,4',6-TETRA-HYDROXYAURONE COMPLEX | | Descriptor: | 3',5'-DIBROMO-2',4,4',6'-TETRAHYDROXY AURONE, TRANSTHYRETIN | | Authors: | Ciszak, E, Cody, V, Luft, J.R. | | Deposit date: | 1992-04-20 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure determination at 2.3-A resolution of human transthyretin-3',5'-dibromo-2',4,4',6-tetrahydroxyaurone complex.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
4LP0
 
 | | Crystal structure of a topoisomerase ATP inhibitor | | Descriptor: | 6'-[(ethylcarbamoyl)amino]-4'-[4-(trifluoromethyl)-1,3-thiazol-2-yl]-3,3'-bipyridine-5-carboxylic acid, Topoisomerase IV subunit B | | Authors: | Basarab, G.S, Manchester, J.I, Bist, S, Boriack-Sjodin, P.A, Dangel, B, Illingsworth, R, Uria-Nickelsen, M, Sherer, B.A, Sriram, S, Eakin, A.E. | | Deposit date: | 2013-07-14 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-to-Hit-to-Lead Discovery of a Novel Pyridylurea Scaffold of ATP Competitive Dual Targeting Type II Topoisomerase Inhibiting Antibacterial Agents.
J.Med.Chem., 56, 2013
|
|
4LPB
 
 | | Crystal structure of a topoisomerase ATPase inhibitor | | Descriptor: | 1-ethyl-3-{5'-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)-4-[4-(trifluoromethyl)-1,3-thiazol-2-yl]-3,3'-bipyridin-6-yl}urea, Topoisomerase IV subunit B | | Authors: | Boriack-Sjodin, A. | | Deposit date: | 2013-07-15 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment-to-Hit-to-Lead Discovery of a Novel Pyridylurea Scaffold of ATP Competitive Dual Targeting Type II Topoisomerase Inhibiting Antibacterial Agents.
J.Med.Chem., 56, 2013
|
|
4ANM
 
 | | Complex of CK2 with a CDC7 inhibitor | | Descriptor: | 8-BROMANYL-2-[[(3S)-3-OXIDANYLPYRROLIDIN-1-YL]METHYL]-3H-[1]BENZOFURO[3,2-D]PYRIMIDIN-4-ONE, CASEIN KINASE II SUBUNIT ALPHA | | Authors: | Stout, T.J. | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Xl413, a Potent and Selective Cdc7 Inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4NRI
 
 | |
8W1D
 
 | | CRYSTAL STRUCTURE OF DPS-LIKE PROTEIN PA4880 FROM PSEUDOMONAS AERUGINOSA (DIMERIC FORM) | | Descriptor: | DPS-LIKE PROTEIN, FE (II) ION | | Authors: | Lovell, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-02-15 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pseudomonas aeruginosa gene PA4880 encodes a Dps-like protein with a Dps fold, bacterioferritin-type ferroxidase centers, and endonuclease activity.
Front Mol Biosci, 11, 2024
|
|
8W1E
 
 | | Crystal Structure of DPS-like protein PA4880 from Pseudomonas aeruginosa (dodecamer) | | Descriptor: | DPS-LIKE PROTEIN, FE (II) ION, SULFATE ION | | Authors: | Lovell, S, Liu, L, Seibold, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-02-15 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Pseudomonas aeruginosa gene PA4880 encodes a Dps-like protein with a Dps fold, bacterioferritin-type ferroxidase centers, and endonuclease activity.
Front Mol Biosci, 11, 2024
|
|
