5GHK
 
 | | Crystal Structure Analysis of Canine serum albumin | | Descriptor: | Serum albumin | | Authors: | Kihira, K, Yamada, K, Kureishi, M, Yokomaku, K, Shinohara, R, Akiyama, M, Komatsu, T. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Artificial Blood for Dogs
Sci Rep, 6, 2016
|
|
5FZI
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with MC3095 | | Descriptor: | 1,2-ETHANEDIOL, 6-oxo-2-[(2-oxo-2-phenylethyl)sulfanyl]-1,6-dihydropyrimidine-5-carboxylic acid, CHLORIDE ION, ... | | Authors: | Nowak, R, Kopec, J, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Rotili, D, Mai, A, Oppermann, U. | | Deposit date: | 2016-03-14 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Mc3095
To be Published
|
|
5FZ3
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment 3,6-Dihydroxybenzonorbornane (N08776b) (ligand modelled based on PANDDA event map) | | Descriptor: | (1R,4S)-1,2,3,4-tetrahydro-1,4-methanonaphthalene-5,8-diol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Pearce, N, Talon, R, Collins, P, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with N08776B
To be Published
|
|
5FMD
 
 | |
5FKS
 
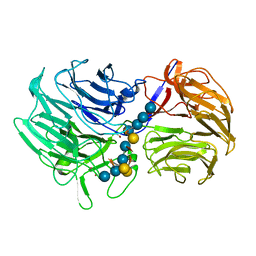 | | Unraveling the first step of xyloglucan degradation by the soil saprophyte Cellvibrio japonicus through the functional and structural characterization of a potent GH74 endo-xyloglucanase | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, ENDO-1,4-BETA-GLUCANASE/XYLOGLUCANASE, ... | | Authors: | Attia, M, Stepper, J, Davies, G.J, Brumer, H. | | Deposit date: | 2015-10-19 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional and Structural Characterization of a Potent Gh74 Endo-Xyloglucanase from the Soil Saprophyte Cellvibrio Japonicus Unravels the First Step of Xyloglucan Degradation.
FEBS J., 283, 2016
|
|
5FMB
 
 | |
5HBV
 
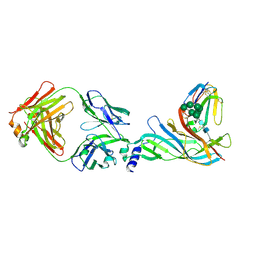 | | Complex structure of Fab35 and mouse nAChR alpha1 | | Descriptor: | Acetylcholine receptor subunit alpha 1, Alpha-bungarotoxin isoform V31, Fab35, ... | | Authors: | Noridomi, K, Watanabe, G, Hansen, M.N, Han, G.W, Chen, L. | | Deposit date: | 2016-01-02 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the molecular mechanisms of myasthenia gravis and their therapeutic implications.
Elife, 6, 2017
|
|
5HYI
 
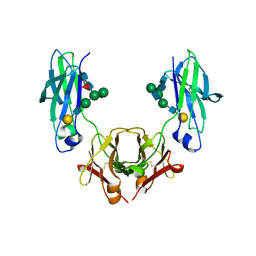 | | Glycosylated, disulfide-linked Hole-Hole Fc fragment | | Descriptor: | Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural differences between glycosylated, disulfide-linked heterodimeric Knob-into-Hole Fc fragment and its homodimeric Knob-Knob and Hole-Hole side products.
Protein Eng. Des. Sel., 30, 2017
|
|
5HO9
 
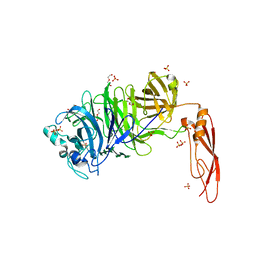 | | Structure of truncated AbnA (domains 1-3), a GH43 arabinanase from Geobacilllus stearothermophilus, in complex with arabinooctaose | | Descriptor: | DI(HYDROXYETHYL)ETHER, Extracellular arabinanase, SULFATE ION, ... | | Authors: | Lansky, S, Salama, R, Azoulai, D, Shwartstien, O, Shoham, Y, Shoham, G. | | Deposit date: | 2016-01-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.845 Å) | | Cite: | Structure of truncated AbnA (domains 1-3), a GH43 arabinanase from Geobacilllus stearothermophilus, in complex with arabinooctaose
To Be Published
|
|
5HBT
 
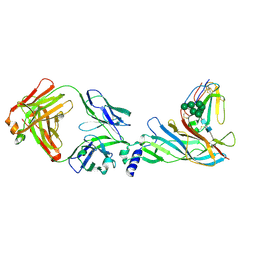 | | Complex structure of Fab35 and human nAChR alpha1 | | Descriptor: | Acetylcholine receptor subunit alpha 1, Alpha-bungarotoxin isoform V31, Fab35, ... | | Authors: | Noridomi, K, Watanabe, G, Hansen, M.N, Han, G.W, Chen, L. | | Deposit date: | 2016-01-02 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural insights into the molecular mechanisms of myasthenia gravis and their therapeutic implications.
Elife, 6, 2017
|
|
7F59
 
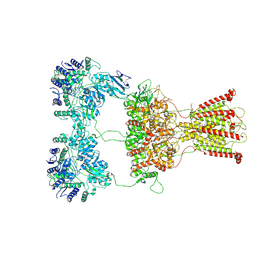 | | DNQX-bound GluK2-1xNeto2 complex | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F56
 
 | | DNQX-bound GluK2-1xNeto2 complex, with asymmetric LBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F5B
 
 | | LBD-TMD focused reconstruction of DNQX-bound GluK2-1xNeto2 complex | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F5A
 
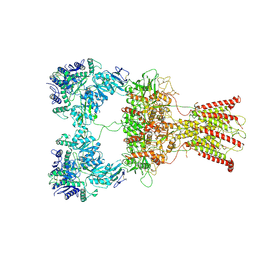 | | DNQX-bound GluK2-2xNeto2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F57
 
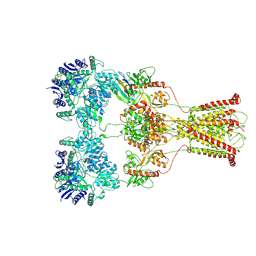 | | Kainate-bound GluK2-1xNeto2 complex, at the desensitized state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7ESV
 
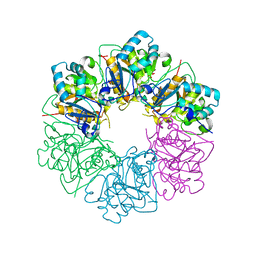 | |
7ESQ
 
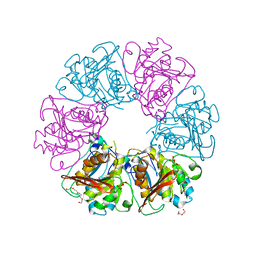 | |
7ESP
 
 | |
7ESO
 
 | |
7FAP
 
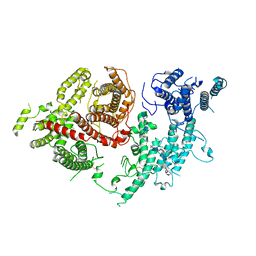 | | Structure of VAR2CSA-CSA 3D7 | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-3)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, Erythrocyte membrane protein 1, PfEMP1 | | Authors: | Wang, L, Wang, Z. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The molecular mechanism of cytoadherence to placental or tumor cells through VAR2CSA from Plasmodium falciparum.
Cell Discov, 7, 2021
|
|
7EY2
 
 | |
7K3B
 
 | | Structure of full-length influenza HA with a head-binding antibody at pH 5.2, conformation C, central helices splay | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Gao, J, Xiang, Y. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7K3A
 
 | | Structure of full-length influenza HA with a head-binding antibody at pH 5.2, conformation B, fusion peptide release | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Gao, J, Xiang, Y. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7K37
 
 | | Structure of full-length influenza HA with a head-binding antibody at pH 7.8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Xiang, Y, Gao, J. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7JGH
 
 | | Cryo-EM structure of P. falciparum VAR2CSA NF54 core in complex with CSA at 3.36 A | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, Erythrocyte membrane protein 1, beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid | | Authors: | Ma, R, Tolia, N.H. | | Deposit date: | 2020-07-19 | | Release date: | 2021-01-13 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis for placental malaria mediated by Plasmodium falciparum VAR2CSA.
Nat Microbiol, 6, 2021
|
|
