8GGL
 
 | | Locally refined cryoEM structure of receptor from beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #4 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GG0
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #6 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GGY
 
 | | Locally refined cryoEM structure of receptor from beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #17 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8AA7
 
 | | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with a heme coordinated fragment | | Descriptor: | 2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, Cytochrome P450 protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-06-30 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with a heme coordinated fragment
To Be Published
|
|
5MZ4
 
 | | Crystal Structure of full-lengh CSFV NS3/4A | | Descriptor: | Genome polyprotein,Genome polyprotein | | Authors: | Tortorici, M.A, Rey, F.A. | | Deposit date: | 2017-01-30 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.048 Å) | | Cite: | A positive-strand RNA virus uses alternative protein-protein interactions within a viral protease/cofactor complex to switch between RNA replication and virion morphogenesis.
PLoS Pathog., 13, 2017
|
|
8GGF
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #20 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8JWY
 
 | | Crystal structure of A2AR-T4L in complex with 2-118 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-azanyl-6-[2-oxidanylidene-1-[[6-(2-oxidanylpropan-2-yl)pyridin-2-yl]methyl]pyridin-4-yl]pyrimidin-4-yl]-2-methyl-benzenecarbonitrile, Adenosine receptor A2a,Endolysin, ... | | Authors: | Weng, Y, Chen, Y, Xu, Y, Song, G. | | Deposit date: | 2023-06-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insight into the dual-antagonistic mechanism of AB928 on adenosine A 2 receptors.
Sci China Life Sci, 67, 2024
|
|
8ABO
 
 | |
8ABS
 
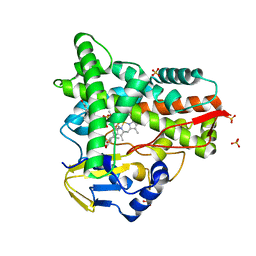 | | Crystal structure of CYP109A2 from Bacillus megaterium bound with testosterone and putative ligand 4,6-dimethyloctanoic acid | | Descriptor: | (4~{S},6~{S})-4,6-dimethyloctanoic acid, Cytochrome P450, HEME B/C, ... | | Authors: | Jozwik, I.K, Rozeboom, H.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Regio- and stereoselective steroid hydroxylation by CYP109A2 from Bacillus megaterium explored by X-ray crystallography and computational modeling.
Febs J., 290, 2023
|
|
8KCZ
 
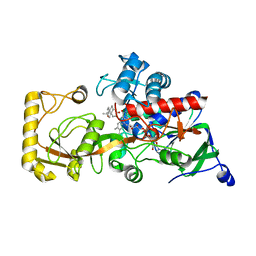 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione | | Descriptor: | 3-ketosteroid dehydrogenase, ANDROSTA-1,4-DIENE-3,17-DIONE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hu, Y.L, Li, X, Cheng, X.Y, Song, S.K, Su, Z.D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione
To Be Published
|
|
8DNS
 
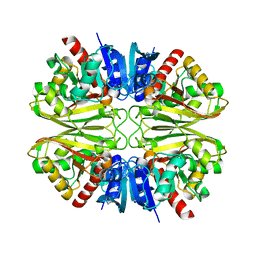 | | Human Brain Glyceraldehyde 3-phosphate dehydrogenase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Tringides, M.L. | | Deposit date: | 2022-07-11 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | A cryo-electron microscopic approach to elucidate protein structures from human brain microsomes.
Life Sci Alliance, 6, 2023
|
|
8AD3
 
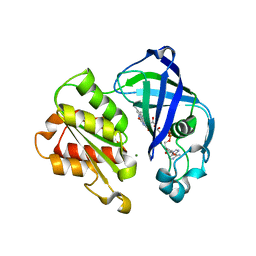 | | X-ray structure of NqrF(129-408)of Vibrio cholerae variant F406A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, Na(+)-translocating NADH-quinone reductase subunit F | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8GG3
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #9 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GGI
 
 | | Locally refined cryoEM structure of receptor from beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #1 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8K1I
 
 | |
1S5J
 
 | | Insight in DNA Replication: The crystal structure of DNA Polymerase B1 from the archaeon Sulfolobus solfataricus | | Descriptor: | DNA polymerase I, MAGNESIUM ION, SULFATE ION | | Authors: | Savino, C, Federici, L, Nastopoulos, V, Johnson, K.A, Pisani, F.M, Rossi, M, Tsernoglou, D. | | Deposit date: | 2004-01-21 | | Release date: | 2004-11-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into DNA replication: the crystal structure of DNA polymerase B1 from the archaeon Sulfolobus solfataricus
Structure, 12, 2004
|
|
8GGT
 
 | | Locally refined cryoEM structure of receptor from beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #12 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
6CP4
 
 | | P450CAM D251N MUTANT | | Descriptor: | CAMPHOR, CYTOCHROME P450CAM, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 1998-05-28 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Understanding the role of the essential Asp251 in cytochrome p450cam using site-directed mutagenesis, crystallography, and kinetic solvent isotope effect.
Biochemistry, 37, 1998
|
|
8ABR
 
 | | Crystal structure of CYP109A2 from Bacillus megaterium bound with putative ligands hexanoic acid and octanoic acid | | Descriptor: | Cytochrome P450, HEME B/C, HEXANOIC ACID, ... | | Authors: | Jozwik, I.K, Rozeboom, H.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Regio- and stereoselective steroid hydroxylation by CYP109A2 from Bacillus megaterium explored by X-ray crystallography and computational modeling.
Febs J., 290, 2023
|
|
8ACK
 
 | |
8ACR
 
 | |
8JWZ
 
 | | Crystal structure of A2AR-T4L in complex with AB928 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-azanyl-6-[1-[[6-(2-oxidanylpropan-2-yl)pyridin-2-yl]methyl]-1,2,3-triazol-4-yl]pyrimidin-4-yl]-2-methyl-benzenecarbonitrile, Adenosine receptor A2a,Endolysin, ... | | Authors: | Weng, Y, Chen, Y, Xu, Y, Song, G. | | Deposit date: | 2023-06-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insight into the dual-antagonistic mechanism of AB928 on adenosine A 2 receptors.
Sci China Life Sci, 67, 2024
|
|
5UJD
 
 | | SbnI from Staphylococcus pseudintermedius | | Descriptor: | FORMIC ACID, Siderophore biosynthesis protein SbnI | | Authors: | Murphy, M.E.P, Verstraete, M.M. | | Deposit date: | 2017-01-17 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The heme-sensitive regulator SbnI has a bifunctional role in staphyloferrin B production by Staphylococcus aureus .
J.Biol.Chem., 294, 2019
|
|
8GG5
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #11 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
1S6L
 
 | | Solution structure of MerB, the Organomercurial Lyase involved in the bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase | | Authors: | Di Lello, P, Benison, G.C, Valafar, H, Pitts, K.E, Summers, A.O, Legault, P, Omichinski, J.G. | | Deposit date: | 2004-01-25 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies reveal a novel protein fold for MerB, the organomercurial lyase involved in the bacterial mercury resistance system.
Biochemistry, 43, 2004
|
|
