2FMY
 
 | |
2H6C
 
 | |
6DT4
 
 | | 1.8 Angstrom Resolution Crystal Structure of cAMP-Regulatory Protein from Yersinia pestis in Complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CHLORIDE ION, Cyclic AMP receptor protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Ritzert, J.T.H, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-15 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Cyclic AMP Receptor Protein Regulates Quorum Sensing and Global Gene Expression in Yersinia pestis during Planktonic Growth and Growth in Biofilms.
Mbio, 10, 2019
|
|
6EO1
 
 | | The electron crystallography structure of the cAMP-bound potassium channel MloK1 (PCO-refined) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Kowal, J, Biyani, N, Chami, M, Scherer, S, Rzepiela, A, Baumgartner, P, Upadhyay, V, Nimigean, C, Stahlberg, H. | | Deposit date: | 2017-10-08 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (4.5 Å) | | Cite: | High-Resolution Cryoelectron Microscopy Structure of the Cyclic Nucleotide-Modulated Potassium Channel MloK1 in a Lipid Bilayer.
Structure, 26, 2018
|
|
6CJU
 
 | | Structure of the SthK cyclic nucleotide-gated potassium channel in complex with cAMP | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, SthK cyclic nucleotide-gated potassium channel | | Authors: | Nimigean, C.M, Rheinberger, J. | | Deposit date: | 2018-02-26 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Ligand discrimination and gating in cyclic nucleotide-gated ion channels from apo and partial agonist-bound cryo-EM structures.
Elife, 7, 2018
|
|
2H6B
 
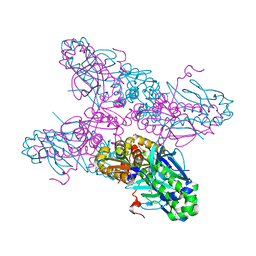 | |
2GZW
 
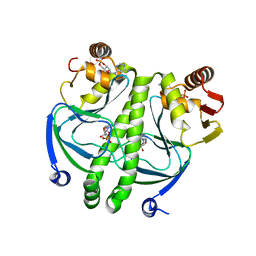 | | Crystal structure of the E.coli CRP-cAMP complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator | | Authors: | Kumarevel, T.S, Tanaka, T, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-12 | | Release date: | 2007-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of activated CRP protein from E coli
To be Published
|
|
5U6P
 
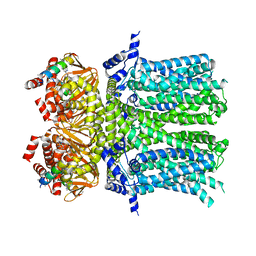 | |
3KCC
 
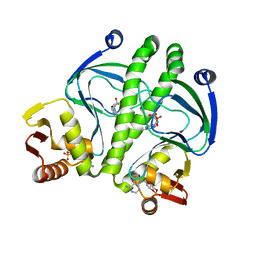 | | Crystal structure of D138L mutant of Catabolite Gene Activator Protein | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator | | Authors: | Tao, W.B, Gao, Z.Q, Zhou, J.H, Dong, Y.H, Yu, S.N. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The 1.6A resolution structure of activated D138L mutant of catabolite gene activator protein with two cAMP bound in each monomer
Int.J.Biol.Macromol., 48, 2011
|
|
8EU3
 
 | | Cryo-EM structure of cGMP bound human CNGA3/CNGB3 channel in GDN, transition state 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel alpha-3, ... | | Authors: | Hu, Z, Zheng, X, Yang, J. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Conformational trajectory of allosteric gating of the human cone photoreceptor cyclic nucleotide-gated channel.
Nat Commun, 14, 2023
|
|
8ETP
 
 | | Cryo-EM structure of cGMP bound closed state of human CNGA3/CNGB3 channel in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel alpha-3, ... | | Authors: | Hu, Z, Zheng, X, Yang, J. | | Deposit date: | 2022-10-17 | | Release date: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Conformational trajectory of allosteric gating of the human cone photoreceptor cyclic nucleotide-gated channel.
Nat Commun, 14, 2023
|
|
8EV9
 
 | |
8EVC
 
 | |
8EV8
 
 | |
7T4V
 
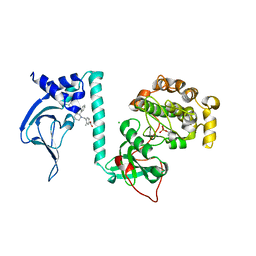 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | 4-({(2S,3S)-3-[(1S)-1-(3,5-dichlorophenyl)-2-hydroxyethoxy]-2-phenylpiperidin-1-yl}methyl)-3-methylbenzoic acid, CHLORIDE ION, cGMP-dependent protein kinase 1 | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
7T4W
 
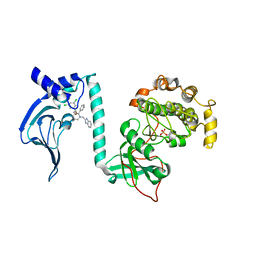 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | (2R)-2-({(2S,3S)-1-[(1H-benzimidazol-2-yl)methyl]-2-phenylpiperidin-3-yl}oxy)-2-(3,5-dichlorophenyl)ethan-1-ol, CHLORIDE ION, cGMP-dependent protein kinase 1 | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
7T4T
 
 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
7T4U
 
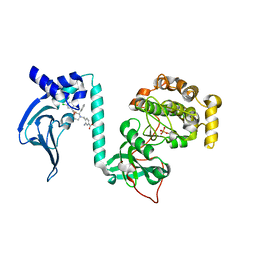 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | 3-amino-4-({(2S,3S)-3-[(1S)-1-(3,5-dichlorophenyl)-2-hydroxyethoxy]-2-phenylpiperidin-1-yl}methyl)benzoic acid, cGMP-dependent protein kinase 1 | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
4F7Z
 
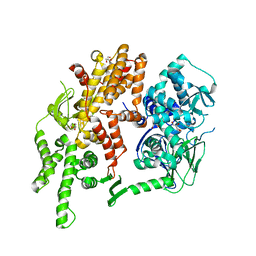 | | Conformational dynamics of exchange protein directly activated by cAMP | | Descriptor: | GLYCEROL, Rap guanine nucleotide exchange factor 4 | | Authors: | White, M.A, Tsalkova, T.N, Mei, F.C, Liu, T, Woods, V.L, Cheng, X. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analyses of a constitutively active mutant of exchange protein directly activated by cAMP.
Plos One, 7, 2012
|
|
3LA7
 
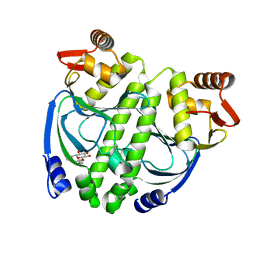 | | Crystal structure of NtcA in apo-form | | Descriptor: | Global nitrogen regulator, octyl beta-D-glucopyranoside | | Authors: | Zhao, M.X, Jiang, Y.L, He, Y.X, Chen, Y.F, Teng, Y.B, Zhang, C.C, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2010-01-06 | | Release date: | 2010-09-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5W5A
 
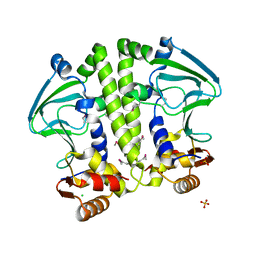 | | Crystal structure of Mycobacterium tuberculosis CRP-FNR family transcription factor Cmr (Rv1675c) | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator Cmr, SULFATE ION | | Authors: | Cheung, J, Cassidy, M, Ginter, C, Ranganathan, S, Pata, D.J, McDonough, K.A. | | Deposit date: | 2017-06-14 | | Release date: | 2017-12-13 | | Last modified: | 2019-01-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Novel structural features drive DNA binding properties of Cmr, a CRP family protein in TB complex mycobacteria.
Nucleic Acids Res., 46, 2018
|
|
5W5B
 
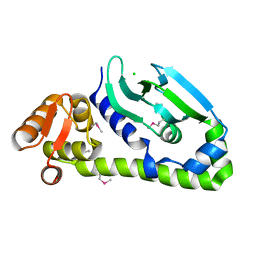 | | Crystal structure of Mycobacterium tuberculosis CRP-FNR family transcription factor Cmr (Rv1675c), truncated construct | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator Cmr | | Authors: | Cheung, J, Cassidy, M, Ginter, C, Ranganathan, S, Pata, D.J, McDonough, K.A. | | Deposit date: | 2017-06-14 | | Release date: | 2017-12-13 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel structural features drive DNA binding properties of Cmr, a CRP family protein in TB complex mycobacteria.
Nucleic Acids Res., 46, 2018
|
|
3LA3
 
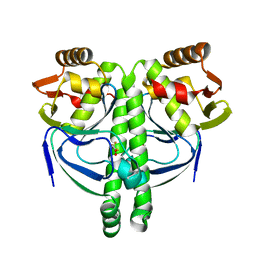 | | Crystal structure of NtcA in complex with 2,2-difluoropentanedioic acid | | Descriptor: | 2,2-difluoropentanedioic acid, Global nitrogen regulator | | Authors: | Zhao, M.X, Jiang, Y.L, He, Y.X, Chen, Y.F, Teng, Y.B, Chen, Y.X, Zhang, C.C, Zhou, C.Z. | | Deposit date: | 2010-01-06 | | Release date: | 2010-07-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4FT8
 
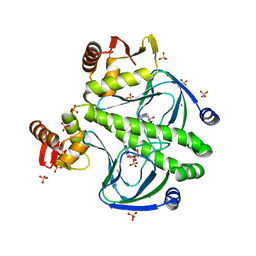 | |
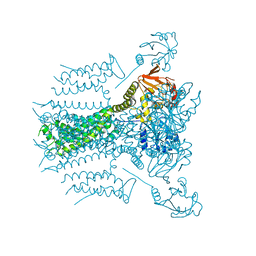
5VA3
 
 | |
