8D7B
 
 | |
5CND
 
 | | Ultrafast dynamics in myoglobin: 3 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
4QI6
 
 | | Cellobiose dehydrogenase from Myriococcum thermophilum, MtCDH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cellobiose dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
8D8E
 
 | |
5MNS
 
 | | Structural and functional characterization of OleP in complex with 6DEB in sodium formate | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Parisi, G, Savino, C, Montemiglio, L.C, Vallone, B. | | Deposit date: | 2016-12-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Substrate-induced conformational change in cytochrome P450 OleP.
FASEB J., 33, 2019
|
|
6BH7
 
 | | Phosphotriesterase variant R18+254S | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-10-30 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphotriesterase variant R18+254S
To Be Published
|
|
8CXI
 
 | | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy | | Descriptor: | A11 heavy chain, A11 light chain, Ankyrin repeat family A protein 2,Envelope E protein, ... | | Authors: | Liu, W, Zhang, X.K, Gong, D.Y, Dai, X.H, Sharma, A, Zhang, T.H, Rey, F, Zhou, Z.H. | | Deposit date: | 2022-05-21 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy
To Be Published
|
|
4QJE
 
 | | 1.85 Angstrom resolution crystal structure of apo betaine aldehyde dehydrogenase (betB) G234S mutant from Staphylococcus aureus (IDP00699) with BME-free sulfinic acid form of Cys289 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Betaine aldehyde dehydrogenase, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-03 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8D5U
 
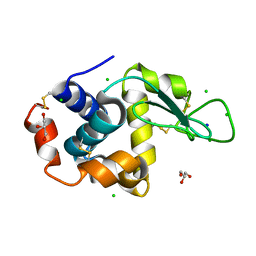 | |
4QOE
 
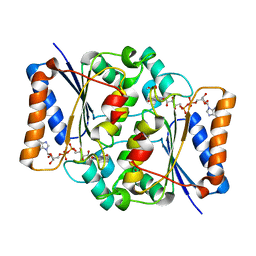 | | The value 'crystal structure of fad quinone reductase 2 at 1.45A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | Deposit date: | 2014-06-20 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The value 'crystal structure of fad quinone reductase 2 at 1.45A
To be Published
|
|
8DGM
 
 | | 14-3-3 epsilon bound to phosphorylated PEAK1 (pT1165) peptide | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein epsilon, Inactive tyrosine-protein kinase PEAK1 | | Authors: | Roy, M.J, Hardy, J.M, Lucet, I.S. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mapping of PEAK pseudokinase interactions identifies 14-3-3 as a molecular switch for PEAK3 signaling.
Nat Commun, 14, 2023
|
|
8D6B
 
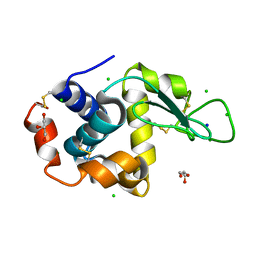 | |
1SM3
 
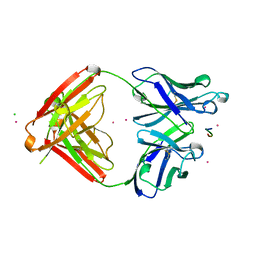 | | CRYSTAL STRUCTURE OF THE TUMOR SPECIFIC ANTIBODY SM3 COMPLEX WITH ITS PEPTIDE EPITOPE | | Descriptor: | CADMIUM ION, CHLORIDE ION, PEPTIDE EPITOPE, ... | | Authors: | Dokurno, P, Bates, P.A, Band, H.A, Stewart, L.M.D, Lally, J.M, Burchell, J.M, Taylor-Papadimitriou, J, Sternberg, M.J.E, Snary, D, Freemont, P.S. | | Deposit date: | 1997-12-23 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure at 1.95 A resolution of the breast tumour-specific antibody SM3 complexed with its peptide epitope reveals novel hypervariable loop recognition.
J.Mol.Biol., 284, 1998
|
|
8D5W
 
 | |
8J0I
 
 | | Aldo-keto reductase KmAKR | | Descriptor: | NADPH-dependent alpha-keto amide reductase, SODIUM ION | | Authors: | Xu, S.Y, Zhou, L, Xu, Y, Wang, Y.J, Zheng, Y.G. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Aldo-keto reductase KmAKR from Kluyveromyces marxianus
To Be Published
|
|
8GCV
 
 | |
8AG6
 
 | |
8CXG
 
 | | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy | | Descriptor: | Ankyrin repeat family A protein 2,Envelope E protein, C8 scFv heavy chain, C8 scFv light chain, ... | | Authors: | Liu, W, Zhang, X.K, Gong, D.Y, Dai, X.H, Sharma, A, Zhang, T.H, Rey, F, Zhou, Z.H. | | Deposit date: | 2022-05-21 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy
To Be Published
|
|
8AJX
 
 | | E. coli NfsA with Fumarate | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, FUMARIC ACID, ... | | Authors: | Day, M.A, Jarrom, D, White, S.A, Hyde, E.I. | | Deposit date: | 2022-07-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Oxygen-insensitive nitroreductase E. coli NfsA, but not NfsB, is inhibited by fumarate.
Proteins, 91, 2023
|
|
8D7C
 
 | |
1AHL
 
 | | ANTHOPLEURIN-A,NMR, 20 STRUCTURES | | Descriptor: | ANTHOPLEURIN-A | | Authors: | Pallaghy, P.K, Scanlon, M.J, Monks, S.A, Norton, R.S. | | Deposit date: | 1994-10-28 | | Release date: | 1995-11-14 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in solution of the polypeptide cardiac stimulant anthopleurin-A.
Biochemistry, 34, 1995
|
|
8D8F
 
 | |
8GCT
 
 | |
4QOG
 
 | | Crystal structure of fad quinone reductase 2 in complex with melatonin at 1.4A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | Deposit date: | 2014-06-20 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of fad quinone reductase 2 in complex with melatonin at 1.4A
To be Published
|
|
8AI7
 
 | | Structure of carbamoylated human butyrylcholinesterase upon reaction with 3-(((2-cycloheptylethyl)(methyl)amino)methyl)-1H-indol-7-yl N,N-dimethylcarbamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[[2-cycloheptylethyl(methyl)amino]methyl]-1~{H}-indol-7-ol, ... | | Authors: | Brazzolotto, X, Meden, A, Knez, D, Gobec, S, Nachon, F. | | Deposit date: | 2022-07-25 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Pseudo-irreversible butyrylcholinesterase inhibitors: Structure-activity relationships, computational and crystallographic study of the N-dialkyl O-arylcarbamate warhead.
Eur.J.Med.Chem., 247, 2023
|
|
