2XDP
 
 | | Crystal structure of the tudor domain of human JMJD2C | | Descriptor: | LYSINE-SPECIFIC DEMETHYLASE 4C, SULFATE ION | | Authors: | Yue, W.W, Gileadi, C, Krojer, T, Weisbach, H, Ugochukwu, E, Daniel, M, Phillips, C, Chaikuad, A, von Delft, F, Allerston, C, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2010-05-06 | | Release date: | 2010-06-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal Structure of the Tudor Domain of Human Jmjd2C
To be Published
|
|
4F1J
 
 | | Crystal structure of the MG2+ loaded VWA domain of plasmodium falciparum trap protein | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Pihlajamaa, T, Knuuti, J, Kajander, T, Sharma, A, Permi, P. | | Deposit date: | 2012-05-07 | | Release date: | 2013-01-30 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure of Plasmodium falciparum TRAP (thrombospondin-related anonymous protein) A domain highlights distinct features in apicomplexan von Willebrand factor A homologues.
Biochem.J., 450, 2013
|
|
4F1U
 
 | | Subatomic resolution structure of a high affinity periplasmic phosphate-binding protein (PfluDING) bound with phosphate at pH 4.5 | | Descriptor: | 1,2-ETHANEDIOL, HYDROGENPHOSPHATE ION, Putative alkaline phosphatase, ... | | Authors: | Liebschner, D, Elias, M, Tawfik, D.S, Moniot, S, Fournier, B, Scott, K, Jelsch, C, Guillot, B, Lecomte, C, Chabriere, E. | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | The molecular basis of phosphate discrimination in arsenate-rich environments.
Nature, 491, 2012
|
|
2XCV
 
 | | Crystal structure of the D52N variant of cytosolic 5'-nucleotidase II in complex with inosine monophosphate and 2,3-bisphosphoglycerate | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, CYTOSOLIC PURINE 5'-NUCLEOTIDASE, GLYCEROL, ... | | Authors: | Wallden, K, Nordlund, P. | | Deposit date: | 2010-04-26 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Allosteric Regulation and Substrate Recognition of Human Cytosolic 5'-Nucleotidase II
J.Mol.Biol., 408, 2011
|
|
2XDA
 
 | | STRUCTURE OF HELICOBACTER PYLORI TYPE II DEHYDROQUINASE IN COMPLEX WITH INHIBITOR COMPOUND (4R,6R,7S)-2-(2-Cyclopropyl)ethyl-4,6,7- trihydroxy-4,5,6,7-tetrahydrobenzo(b)thiophene-4-carboxylic acid | | Descriptor: | (4R,6R,7S)-2-(2-CYCLOPROPYLETHYL)-4,6,7-TRIHYDROXY-4,5,6,7-TETRAHYDRO-1-BENZOTHIOPHENE-4-CARBOXYLIC ACID, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Paz, S, Tizon, L, Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, van Raaij, M.J, Lamb, H, Hawkins, A.R, Castedo, L, Gonzalez-Bello, C. | | Deposit date: | 2010-04-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tetrahydrobenzothiophene derivatives: conformationally restricted inhibitors of type II dehydroquinase.
ChemMedChem, 6, 2011
|
|
2XOF
 
 | | Ribonucleotide reductase Y122NO2Y modified R2 subunit of E. coli | | Descriptor: | MU-OXO-DIIRON, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT BETA | | Authors: | Yokoyama, K, Uhlin, U, Stubbe, J. | | Deposit date: | 2010-08-15 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Hot Oxidant, 3-No(2)Y(122) Radical, Unmasks Conformational Gating in Ribonucleotide Reductase.
J.Am.Chem.Soc., 132, 2010
|
|
1HLS
 
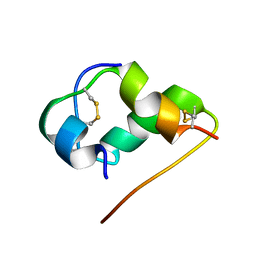 | | NMR STRUCTURE OF THE HUMAN INSULIN-HIS(B16) | | Descriptor: | INSULIN | | Authors: | Ludvigsen, S, Kaarsholm, N.C. | | Deposit date: | 1995-06-28 | | Release date: | 1995-09-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of an engineered biologically potent insulin monomer, B16 Tyr-->His, as determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 33, 1994
|
|
1I02
 
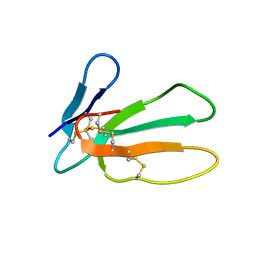 | |
8F6N
 
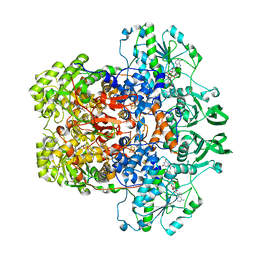 | | Dihydropyrimidine Dehydrogenase (DPD) C671S Mutant Soaked with Thymine Quasi-Anaerobically | | Descriptor: | Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kaley, N, Smith, M, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2022-11-16 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Mammalian dihydropyrimidine dehydrogenase: Added mechanistic details from transient-state analysis of charge transfer complexes.
Arch.Biochem.Biophys., 736, 2023
|
|
4F61
 
 | | Tubulin:Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gigant, B, Mignot, I, Knossow, M. | | Deposit date: | 2012-05-14 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.17 Å) | | Cite: | Design and characterization of modular scaffolds for tubulin assembly.
J.Biol.Chem., 287, 2012
|
|
1IB0
 
 | |
8EMH
 
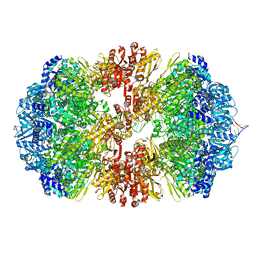 | |
4FE3
 
 | |
1HQR
 
 | | CRYSTAL STRUCTURE OF A SUPERANTIGEN BOUND TO THE HIGH-AFFINITY, ZINC-DEPENDENT SITE ON MHC CLASS II | | Descriptor: | HLA-DR ALPHA CHAIN, HLA-DR BETA CHAIN, MYELIN BASIC PROTEIN, ... | | Authors: | Li, Y, Li, H, Dimasi, N, Schlievert, P, Mariuzza, R. | | Deposit date: | 2000-12-19 | | Release date: | 2001-01-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a superantigen bound to the high-affinity, zinc-dependent site on MHC class II.
Immunity, 14, 2001
|
|
4F1V
 
 | | Subatomic resolution structure of a high affinity periplasmic phosphate-binding protein (PfluDING) bound with phosphate at pH 8.5 | | Descriptor: | HYDROGENPHOSPHATE ION, Putative alkaline phosphatase, SULFATE ION | | Authors: | Liebschner, D, Elias, M, Tawfik, D.S, Moniot, S, Fournier, B, Scott, K, Jelsch, C, Guillot, B, Lecomte, C, Chabriere, E. | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | The molecular basis of phosphate discrimination in arsenate-rich environments.
Nature, 491, 2012
|
|
1HR8
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant Complexed with Cytochrome C Oxidase IV Signal Peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CYTOCHROME C OXIDASE POLYPEPTIDE IV, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
2YFY
 
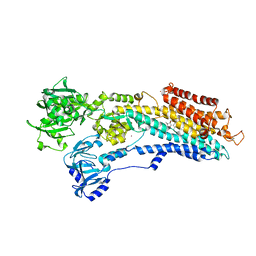 | | SERCA in the HnE2 State Complexed With Debutanoyl Thapsigargin | | Descriptor: | DEBUTANOYL THAPSIGARGIN, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Sonntag, Y, Musgaard, M, Olesen, C, Schiott, B, Moller, J.V, Nissen, P, Thogersen, L. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mutual Adaptation of a Membrane Protein and its Lipid Bilayer During Conformational Changes.
Nat.Commun., 2, 2011
|
|
2XD9
 
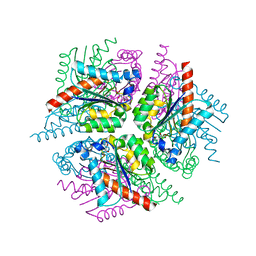 | | STRUCTURE OF HELICOBACTER PYLORI TYPE II DEHYDROQUINASE IN COMPLEX WITH INHIBITOR COMPOUND (4R,6R,7S)-4,6,7-Trihydroxy-2-((E)-prop-1- enyl)-4,5,6,7-tetrahydrobenzo(b)thiophene-4-carboxylic acid | | Descriptor: | (4R,6R,7S)-4,6,7-TRIHYDROXY-2-[(1E)-PROP-1-EN-1-YL]-4,5,6,7-TETRAHYDRO-1-BENZOTHIOPHENE-4-CARBOXYLIC ACID, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Paz, S, Tizon, L, Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, van Raaij, M.J, Lamb, H, Hawkins, A.R, Lapthorn, A.J, Castedo, L, Gonzalez-Bello, C. | | Deposit date: | 2010-04-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tetrahydrobenzothiophene derivatives: conformationally restricted inhibitors of type II dehydroquinase.
ChemMedChem, 6, 2011
|
|
1HVZ
 
 | |
2XKN
 
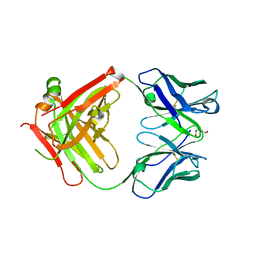 | | Crystal structure of the Fab fragment of the anti-EGFR antibody 7A7 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ANTI-EGFR ANTIBODY 7A7 | | Authors: | Talavera, A, Mackenzie, J, Friemann, R, Krengel, U. | | Deposit date: | 2010-07-09 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Fab Fragment of the Anti-Murine Egfr Antibody 7A7 and Exploration of its Receptor Binding Site.
Mol.Immunol., 48, 2011
|
|
2XML
 
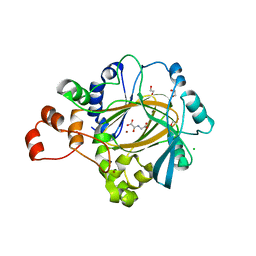 | | Crystal structure of human JMJD2C catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, LYSINE-SPECIFIC DEMETHYLASE 4C, ... | | Authors: | Yue, W.W, Gileadi, C, Krojer, T, Pike, A.C.W, von Delft, F, Ng, S, Carpenter, L, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2010-07-28 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Evolutionary Basis for the Dual Substrate Selectivity of Human Kdm4 Histone Demethylase Family.
J.Biol.Chem., 286, 2011
|
|
4X3H
 
 | | CRYSTAL STRUCTURE OF ARC N-LOBE COMPLEXED WITH STARGAZIN PEPTIDE | | Descriptor: | Activity-regulated cytoskeleton-associated protein, VOLTAGE-DEPENDENT CALCIUM CHANNEL GAMMA-2 SUBUNIT | | Authors: | zhang, W, ward, m, leahy, d, worley, p. | | Deposit date: | 2014-11-30 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis of arc binding to synaptic proteins: implications for cognitive disease.
Neuron, 86, 2015
|
|
2XS4
 
 | | Structure of karilysin catalytic MMP domain in complex with magnesium | | Descriptor: | CHLORIDE ION, KARILYSIN PROTEASE, MAGNESIUM ION, ... | | Authors: | Cerda-Costa, N, Guevara, T, Karim, A.Y, Ksiazek, M, Nguyen, K.-A, Arolas, J.L, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of the Catalytic Domain of Tannerella Forsythia Karilysin Reveals It is a Bacterial Xenologue of Animal Matrix Metalloproteinases.
Mol.Microbiol., 79, 2011
|
|
1IAJ
 
 | | CRYSTAL STRUCTURE OF THE ATYPICAL PROTEIN KINASE DOMAIN OF A TRP CA-CHANNEL, CHAK (APO) | | Descriptor: | TRANSIENT RECEPTOR POTENTIAL-RELATED PROTEIN, ZINC ION | | Authors: | Yamaguchi, H, Matsushita, M, Nairn, A.C, Kuriyan, J. | | Deposit date: | 2001-03-22 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity.
Mol.Cell, 7, 2001
|
|
2Y0R
 
 | | Structural basis for the allosteric interference of myosin function by mutants G680A and G680V of Dictyostelium myosin-2 | | Descriptor: | MYOSIN-2 HEAVY CHAIN | | Authors: | Preller, M, Bauer, S, Adamek, N, Fujita-Becker, S, Fedorov, R, Geeves, M.A, Manstein, D.J. | | Deposit date: | 2010-12-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Allosteric Interference of Myosin Function by Reactive Thiol Region Mutations G680A and G680V.
J.Biol.Chem., 286, 2011
|
|
