7KFU
 
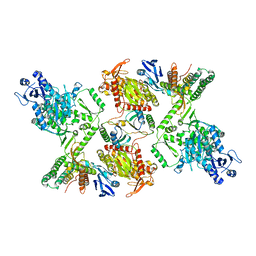 | | Cas6-RT-Cas1--Cas2 complex | | Descriptor: | Cas2, Cas6-RT-Cas1 | | Authors: | Hoel, C.M, Wang, J.Y, Doudna, J.A, Brohawn, S.G. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural coordination between active sites of a CRISPR reverse transcriptase-integrase complex.
Nat Commun, 12, 2021
|
|
7KFT
 
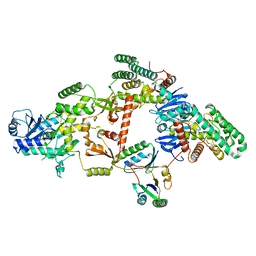 | | Partial Cas6-RT-Cas1--Cas2 complex | | Descriptor: | Cas2, Cas6-RT-Cas1 | | Authors: | Hoel, C.M, Wang, J.Y, Doudna, J.A, Brohawn, S.G. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural coordination between active sites of a CRISPR reverse transcriptase-integrase complex.
Nat Commun, 12, 2021
|
|
7KNR
 
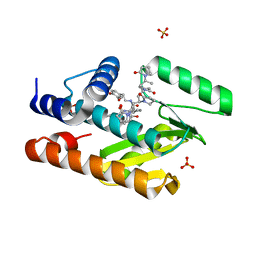 | | The crystal structure of the I38T mutant PA endonuclease (2009/H1N1/California) in complex with SJ000988558 | | Descriptor: | 2-(2,6-difluorophenyl)-5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, White, S.W. | | Deposit date: | 2020-11-05 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The crystal structure of the I38T mutant PA endonuclease (2009/H1N1/California) in complex with SJ000988558
To Be Published
|
|
7KNY
 
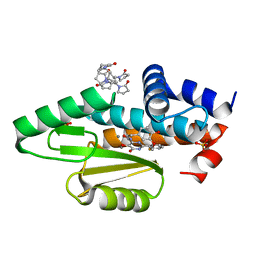 | | The crystal structure of the I38T mutant PA endonuclease (2009/H1N1/California) in complex with SJ000988528 | | Descriptor: | 2-(2-fluorophenyl)-5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, White, S.W. | | Deposit date: | 2020-11-06 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The crystal structure of the I38T mutant PA endonuclease (2009/H1N1/California) in complex with SJ000988528
To Be Published
|
|
7KOP
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease I38T mutant in complex with SJ000988539 | | Descriptor: | 2-(2-fluorophenyl)-5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Seetharaman, J, White, S.W. | | Deposit date: | 2020-11-09 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The crystal structure of the 2009/H1N1/California PA endonuclease I38T mutant in complex with SJ000988539
To Be Published
|
|
7LP8
 
 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000983476 | | Descriptor: | (phenylmethyl) (2~{S})-2-[5-oxidanyl-6-oxidanylidene-4-(2-pyridin-4-ylethylcarbamoyl)-1~{H}-pyrimidin-2-yl]pyrrolidine-1-carboxylate, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000983476
To Be Published
|
|
7LP7
 
 | | The crystal structure of the wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000983476 | | Descriptor: | (phenylmethyl) (2~{S})-2-[5-oxidanyl-6-oxidanylidene-4-(2-pyridin-4-ylethylcarbamoyl)-1~{H}-pyrimidin-2-yl]pyrrolidine-1-carboxylate, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000983476
To Be Published
|
|
7PNY
 
 | | Assembly intermediate of human mitochondrial ribosome small subunit without mS37 in complex with RBFA and METTL15 conformation b | | Descriptor: | 12S mitochondrial rRNA, 12S rRNA N4-methylcytidine (m4C) methyltransferase, 28S ribosomal protein S10, ... | | Authors: | Itoh, Y, Khawaja, A, Rorbach, J, Amunts, A. | | Deposit date: | 2021-09-08 | | Release date: | 2022-06-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Mechanism of mitoribosomal small subunit biogenesis and preinitiation.
Nature, 606, 2022
|
|
7SR6
 
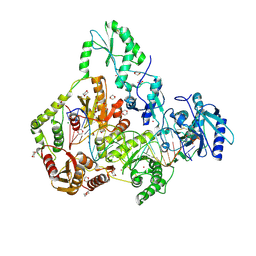 | | Human Endogenous Retrovirus (HERV-K) reverse transcriptase ternary complex with dsDNA template Primer and dNTP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ... | | Authors: | Baldwin, E.T, Nichols, C. | | Deposit date: | 2021-11-08 | | Release date: | 2022-07-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Human endogenous retrovirus-K (HERV-K) reverse transcriptase (RT) structure and biochemistry reveals remarkable similarities to HIV-1 RT and opportunities for HERV-K-specific inhibition.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PNZ
 
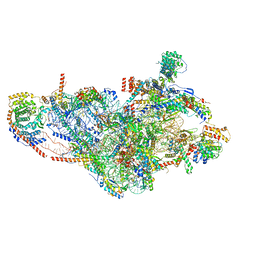 | | Assembly intermediate of human mitochondrial ribosome small subunit without mS37 in complex with RBFA and METTL15 conformation c | | Descriptor: | 12S mitochondrial rRNA, 12S rRNA N4-methylcytidine (m4C) methyltransferase, 28S ribosomal protein S10, ... | | Authors: | Itoh, Y, Khawaja, A, Rorbach, J, Amunts, A. | | Deposit date: | 2021-09-08 | | Release date: | 2022-06-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Mechanism of mitoribosomal small subunit biogenesis and preinitiation.
Nature, 606, 2022
|
|
7PNV
 
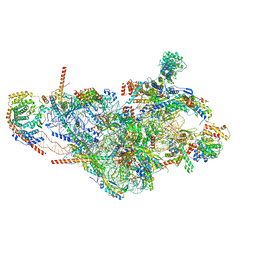 | | Assembly intermediate of mouse mitochondrial ribosome small subunit without mS37 in complex with RbfA and Mettl15 | | Descriptor: | 12S mitochondrial rRNA, 12S rRNA N4-methylcytidine methyltransferase, 28S ribosomal protein S10, ... | | Authors: | Itoh, Y, Khawaja, A, Laptev, I, Sergiev, P, Rorbach, J, Amunts, A. | | Deposit date: | 2021-09-08 | | Release date: | 2022-06-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Mechanism of mitoribosomal small subunit biogenesis and preinitiation.
Nature, 606, 2022
|
|
3IWN
 
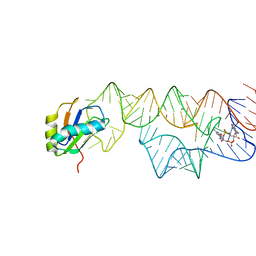 | | Co-crystal structure of a bacterial c-di-GMP riboswitch | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), C-di-GMP riboswitch, U1 small nuclear ribonucleoprotein A | | Authors: | Kulshina, N, Baird, N.J, Ferre-D'Amare, A.R. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6CZR
 
 | | The structure of amicetin bound to the 70S ribosome | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Eiler, D.R, Steitz, T.A, Looper, R.E, Serrano, C.M, Kannareddy, H.R, Koch, M.R, Barrows, L.R, Testa, C.A, Sebahar, P.R. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Unifying the Aminohexopyranose- and Peptidyl-Nucleoside Antibiotics: Implications for Antibiotic Design
Angew.Chem.Int.Ed.Engl., 132, 2020
|
|
3ODR
 
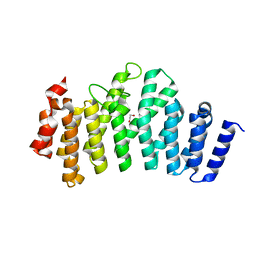 | |
7P2E
 
 | | Human mitochondrial ribosome small subunit in complex with IF3, GMPPMP and streptomycin | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Itoh, Y, Khawaja, A, Singh, V, Rorbach, J, Amunts, A. | | Deposit date: | 2021-07-05 | | Release date: | 2022-07-27 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure of the mitoribosomal small subunit with streptomycin reveals Fe-S clusters and physiological molecules.
Elife, 11, 2022
|
|
3RNT
 
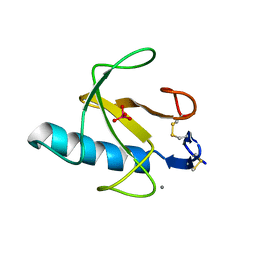 | | CRYSTAL STRUCTURE OF GUANOSINE-FREE RIBONUCLEASE T1, COMPLEXED WITH VANADATE(V), SUGGESTS CONFORMATIONAL CHANGE UPON SUBSTRATE BINDING | | Descriptor: | CALCIUM ION, RIBONUCLEASE T1, VANADATE ION | | Authors: | Kostrewa, D, Choe, H.-W, Heinemann, U, Saenger, W. | | Deposit date: | 1989-05-31 | | Release date: | 1989-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of guanosine-free ribonuclease T1, complexed with vanadate (V), suggests conformational change upon substrate binding.
Biochemistry, 28, 1989
|
|
5E7J
 
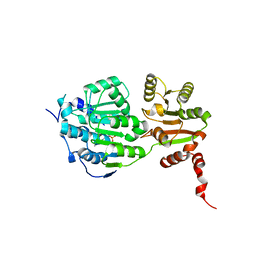 | |
3ODS
 
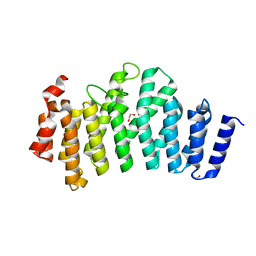 | |
5VHE
 
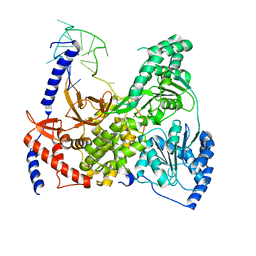 | | DHX36 in complex with the c-Myc G-quadruplex | | Descriptor: | DEAH (Asp-Glu-Ala-His) box polypeptide 36, DNA (5'-D(*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*TP*TP*TP*TP*TP*T)-3'), POTASSIUM ION | | Authors: | Chen, M, Ferre-D'Amare, A. | | Deposit date: | 2017-04-13 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.793 Å) | | Cite: | Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36.
Nature, 558, 2018
|
|
8U3B
 
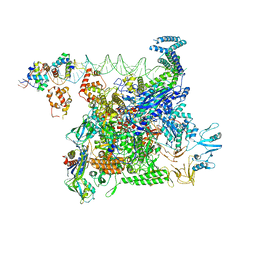 | | Cryo-EM structure of E. coli NarL-transcription activation complex at 3.2A | | Descriptor: | DNA (69-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Kompaniiets, D, Wang, D. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for transcription activation by the nitrate-responsive regulator NarL.
Nucleic Acids Res., 52, 2024
|
|
3KU0
 
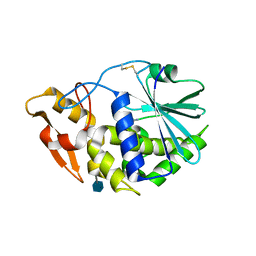 | | Structure of GAP31 with adenine at its binding pocket | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Ribosome-inactivating protein gelonin | | Authors: | Kong, X.-P. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new activity of anti-HIV and anti-tumor protein GAP31: DNA adenosine glycosidase--structural and modeling insight into its functions.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
5E7I
 
 | |
3KTZ
 
 | | Structure of GAP31 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome-inactivating protein gelonin | | Authors: | Kong, X.-P. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new activity of anti-HIV and anti-tumor protein GAP31: DNA adenosine glycosidase--structural and modeling insight into its functions.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
1EAH
 
 | | PV2L COMPLEXED WITH ANTIVIRAL AGENT SCH48973 | | Descriptor: | 1[2-CHLORO-4-METHOXY-PHENYL-OXYMETHYL]-4-[2,6-DICHLORO-PHENYL-OXYMETHYL]-BENZENE, MYRISTIC ACID, POLIOVIRUS TYPE 2 COAT PROTEINS VP1 TO VP4 | | Authors: | Lentz, K, Arnold, E. | | Deposit date: | 1997-07-22 | | Release date: | 1998-09-16 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of poliovirus type 2 Lansing complexed with antiviral agent SCH48973: comparison of the structural and biological properties of three poliovirus serotypes.
Structure, 5, 1997
|
|
1N0O
 
 | | NMR Structure of d(CCAAGGXCTTGGG), X is a 3'-phosphoglycolate, 5'-phosphate gapped lesion, 10 structures | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 5'-D(*CP*CP*AP*AP*GP*G)-3', 5'-D(*CP*CP*CP*AP*AP*GP*GP*CP*CP*TP*TP*GP*G)-3', ... | | Authors: | Junker, H.-D, Hoehn, S.T, Bunt, R.C, Marathius, V, Chen, J, Turner, C.J, Stubbe, J. | | Deposit date: | 2002-10-14 | | Release date: | 2003-01-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Synthesis, Characterization, and Solution Structure of Tethered Oligonucleotides Containing an Internal 3'-Phosphoglycolate, 5'-Phosphate Gapped Lesion
Nucleic Acids Res., 30, 2002
|
|
