8OS3
 
 | | Crystal structure of the titin domain Fn3-11 | | Descriptor: | Titin | | Authors: | Nikoopour, R, Rees, M, Gautel, M. | | Deposit date: | 2023-04-17 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
8SIL
 
 | | Lysozyme crystallized in cyclic olefin copolymer-based microfluidic chips | | Descriptor: | Lysozyme C | | Authors: | Liu, Z, Gu, K, Shelby, M.L, Gilbile, D, Lyubimov, A.Y, Russi, S, Cohen, A.E, Coleman, M.A, Frank, M, Kuhl, T.L, Botha, S, Poitevin, F, Sierra, R.G. | | Deposit date: | 2023-04-16 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A user-friendly plug-and-play cyclic olefin copolymer-based microfluidic chip for room-temperature, fixed-target serial crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8ORM
 
 | | Cryo-EM structure of CAK-THZ1 | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-04-14 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8J22
 
 | | Cryo-EM structure of FFAR2 complex bound with TUG-1375 | | Descriptor: | (2R,4R)-2-(2-chlorophenyl)-3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]carbonyl-1,3-thiazolidine-4-carboxylic acid, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J20
 
 | | Cryo-EM structure of FFAR3 bound with valeric acid and AR420626 | | Descriptor: | (4R)-N-[2,5-bis(chloranyl)phenyl]-4-(furan-2-yl)-2-methyl-5-oxidanylidene-4,6,7,8-tetrahydro-1H-quinoline-3-carboxamide, Free fatty acid receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J24
 
 | | Cryo-EM structure of FFAR2 complex bound with acetic acid | | Descriptor: | ACETATE ION, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Tang, W, Sun, X, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J21
 
 | | Cryo-EM structure of FFAR3 complex bound with butyrate acid | | Descriptor: | Free fatty acid receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J23
 
 | | Cryo-EM structure of FFAR2 complex in apo state | | Descriptor: | Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition and activation mechanism of short chain fatty acid receptors FFAR2 and FFAR3
To Be Published
|
|
8J2M
 
 | | The truncated rice Na+/H+ antiporter SOS1 (1-976) in a constitutively active state | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Na+/H+ antiporter | | Authors: | Zhang, X.Y, Tang, L.H, Zhang, C.R, Nie, J.W. | | Deposit date: | 2023-04-14 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and activation mechanism of the rice Salt Overly Sensitive 1 (SOS1) Na + /H + antiporter.
Nat.Plants, 9, 2023
|
|
8ORL
 
 | |
8SH7
 
 | |
8OR4
 
 | | Partially dissociated CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, Cyclin-dependent kinase 2, ... | | Authors: | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | Deposit date: | 2023-04-13 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
8ORD
 
 | | Cryo-EM map of zebrafish cardiac F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha 1b, ... | | Authors: | Bradshaw, M, Squire, J.M, Morris, E, Atkinson, G, Richardson, B, Lees, J, Paul, D.M. | | Deposit date: | 2023-04-13 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Zebrafish as a model for cardiac disease; Cryo-EM structure of native cardiac thin filaments from Danio Rerio.
J.Muscle Res.Cell.Motil., 44, 2023
|
|
8J19
 
 | | Cryo-EM structure of the LY237-bound GPR84 receptor-Gi complex | | Descriptor: | 6-nonylpyridine-2,4-diol, Antibody fragment ScFv16, G-protein coupled receptor 84, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
8J1A
 
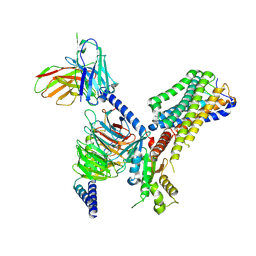 | | Cryo-EM structure of the GPR84 receptor-Gi complex with no ligand modeled | | Descriptor: | Antibody fragment ScFv16, G-protein coupled receptor 84, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
8J18
 
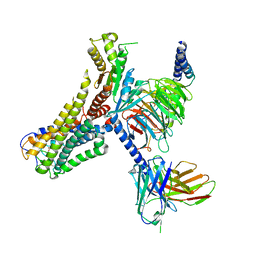 | | Cryo-EM structure of the 3-OH-C12-bound GPR84 receptor-Gi complex | | Descriptor: | (3R)-3-HYDROXYDODECANOIC ACID, Antibody fragment ScFv16, G-protein coupled receptor 84, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
8J1F
 
 | | GSK101 bound state of mTRPV4 | | Descriptor: | N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, Transient receptor potential cation channel subfamily V member 4 | | Authors: | Zhen, W.X, Yang, F. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
8J1H
 
 | | Agonist1 and Ruthenium Red bound state of mTRPV4 | | Descriptor: | 8-fluoranyl-3-[4-(4-fluoranylphenoxy)phenyl]-2-(4-methylpiperazin-1-yl)quinazolin-4-one, Transient receptor potential cation channel subfamily V member 4 | | Authors: | Zhen, W.X, Yang, F. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
8J1D
 
 | | Cryo-EM structure of apo state mTRPV4 | | Descriptor: | Transient receptor potential cation channel subfamily V member 4 | | Authors: | Zhen, W.X, Yang, F. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
8J1B
 
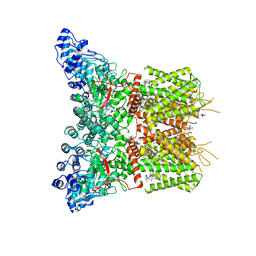 | | GSK101 and Ruthenium Red bound state of mTRPV4 | | Descriptor: | N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, Transient receptor potential cation channel subfamily V member 4, ruthenium(6+) azanide pentaamino(oxido)ruthenium (1/4/2) | | Authors: | Zhen, W.X, Yang, F. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
8SGH
 
 | |
8OR0
 
 | | CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, Cyclin-dependent kinase 2, ... | | Authors: | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
8OQC
 
 | | Dirhodium tetraacetate/ribonuclease A adduct in the P3221 space group (1 h soaking) | | Descriptor: | CHLORIDE ION, FORMIC ACID, Ribonuclease pancreatic, ... | | Authors: | Loreto, D, Merlino, A, Maity, B, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cross-Linked Crystals of Dirhodium Tetraacetate/RNase A Adduct Can Be Used as Heterogeneous Catalysts.
Inorg.Chem., 62, 2023
|
|
8OQG
 
 | | Cross-linked crystal of dirhodium tetraacetate/ribonuclease A adduct in the P3221 space group (high temperature data collection) | | Descriptor: | CHLORIDE ION, FORMIC ACID, Ribonuclease pancreatic, ... | | Authors: | Loreto, D, Merlino, A, Maity, B, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cross-Linked Crystals of Dirhodium Tetraacetate/RNase A Adduct Can Be Used as Heterogeneous Catalysts.
Inorg.Chem., 62, 2023
|
|
8OQD
 
 | | Dirhodium tetraacetate/ribonuclease A adduct in the P3221 space group (1 h soaking) | | Descriptor: | CHLORIDE ION, FORMIC ACID, Ribonuclease pancreatic, ... | | Authors: | Loreto, D, Merlino, A, Maity, B, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Cross-Linked Crystals of Dirhodium Tetraacetate/RNase A Adduct Can Be Used as Heterogeneous Catalysts.
Inorg.Chem., 62, 2023
|
|
