3IFV
 
 | | Crystal structure of the Haloferax volcanii proliferating cell nuclear antigen | | Descriptor: | PCNA, SODIUM ION | | Authors: | Winter, J.A, Christofi, P, Morroll, S, Bunting, K.A. | | Deposit date: | 2009-07-26 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Haloferax volcanii proliferating cell nuclear antigen reveals unique surface charge characteristics due to halophilic adaptation
Bmc Struct.Biol., 9, 2009
|
|
2BWU
 
 | | Asp271Ala Escherichia coli Aminopeptidase P | | Descriptor: | AMINOPEPTIDASE P, CITRATE ANION, MAGNESIUM ION, ... | | Authors: | Graham, S.C, Guss, J.M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
5EMG
 
 | | Crystal structures of PNA p(GCTGCTGC)2 duplex containing T-T mismatches | | Descriptor: | CHLORIDE ION, GPN-CPN-TPN-GPN-CPN-TPN-GPN-CPN, SODIUM ION | | Authors: | Kiliszek, A, Banaszak, K, Dauter, Z, Rypniewski, W. | | Deposit date: | 2015-11-06 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | The first crystal structures of RNA-PNA duplexes and a PNA-PNA duplex containing mismatches-toward anti-sense therapy against TREDs.
Nucleic Acids Res., 44, 2016
|
|
6WMV
 
 | | Structure of a phosphatidylinositol-phosphate synthase (PIPS) from Mycobacterium kansasii with evidence of substrate binding | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,3',3''-phosphanetriyltripropanoic acid, AfCTD-Phosphatidylinositol-phosphate synthase (PIPS) fusion, ... | | Authors: | Belcher Dufrisne, M, Jorge, C.D, Timoteo, C.G, Petrou, V.I, Ashraf, K.U, Banerjee, S, Clarke, O.B, Santos, H, Mancia, F. | | Deposit date: | 2020-04-21 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.142 Å) | | Cite: | Structural and Functional Characterization of Phosphatidylinositol-Phosphate Biosynthesis in Mycobacteria.
J.Mol.Biol., 432, 2020
|
|
6NMF
 
 | | SFX structure of reduced cytochrome c oxidase at room temperature | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Rousseau, D.L, Yeh, S.-R, Ishigami, I. | | Deposit date: | 2019-01-10 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Snapshot of an oxygen intermediate in the catalytic reaction of cytochromecoxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NMP
 
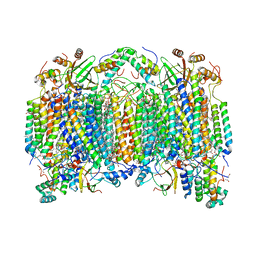 | | SFX structure of oxidized cytochrome c oxidase at room temperature | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Rousseau, D.L, Yeh, S.-R, Ishigami, I. | | Deposit date: | 2019-01-11 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Snapshot of an oxygen intermediate in the catalytic reaction of cytochromecoxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6H6K
 
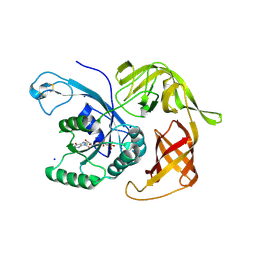 | | The structure of the FKR mutant of the archaeal translation initiation factor 2 gamma subunit in complex with GDPCP, obtained in the absence of magnesium salts in the crystallization solution. | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, SODIUM ION, ... | | Authors: | Nikonov, O, Kravchenko, O, Nevskaya, N, Stolboushkina, E, Gabdulkhakov, A, Garber, M, Nikonov, S. | | Deposit date: | 2018-07-27 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The third structural switch in the archaeal translation initiation factor 2 (aIF2) molecule and its possible role in the initiation of GTP hydrolysis and the removal of aIF2 from the ribosome.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5B2O
 
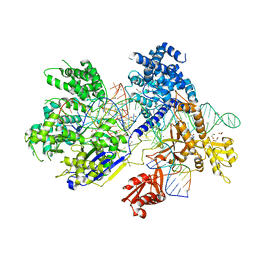 | | Crystal structure of Francisella novicida Cas9 in complex with sgRNA and target DNA (TGG PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Hirano, H, Nishimasu, H, Nakane, T, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-01 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Engineering of Francisella novicida Cas9
Cell, 164, 2016
|
|
5AXR
 
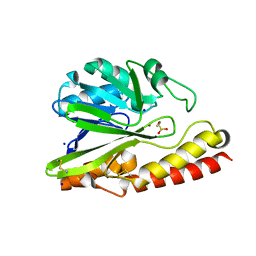 | |
5BJY
 
 | | x-ray structure of the PglF 4,5-dehydratase from campylobacter jejuni, variant M405Y, in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
5IIZ
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F0 | | Descriptor: | Bacterioferritin comigratory protein, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-01 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
6NXS
 
 | | Crystal structure of Arabidopsis thaliana cytosolic triosephosphate isomerase C218Y mutant | | Descriptor: | SODIUM ION, Triosephosphate isomerase, cytosolic | | Authors: | Jimenez-Sandoval, P, Diaz-Quezada, C, Torres-Larios, A, Brieba, L.G. | | Deposit date: | 2019-02-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural basis for the modulation of plant cytosolic triosephosphate isomerase activity by mimicry of redox-based modifications.
Plant J., 99, 2019
|
|
5B2P
 
 | | Crystal structure of Francisella novicida Cas9 in complex with sgRNA and target DNA (TGA PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Hirano, H, Nishimasu, H, Nakane, T, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-01 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Engineering of Francisella novicida Cas9
Cell, 164, 2016
|
|
6O51
 
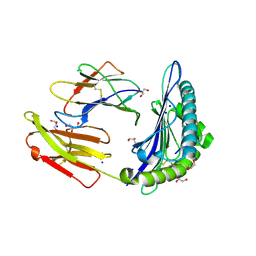 | | Structure of HLA-A2:01 with peptide MM90 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Ying, G, Bitra, A, Zajonc, D.M. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Anin silico-in vitroPipeline Identifying an HLA-A*02:01+KRAS G12V+Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients.
Front Immunol, 10, 2019
|
|
2IZC
 
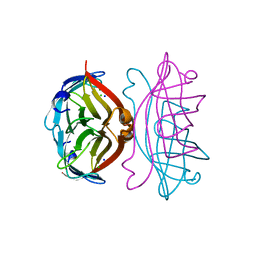 | | APOSTREPTAVIDIN PH 2.0 I222 COMPLEX | | Descriptor: | CHLORIDE ION, SODIUM ION, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
5C4W
 
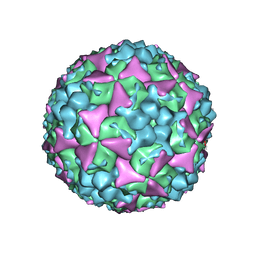 | | Crystal structure of coxsackievirus A16 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SODIUM ION, ... | | Authors: | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
6OME
 
 | |
6O6R
 
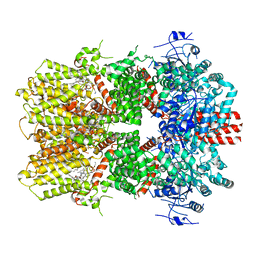 | | Structure of the TRPM8 cold receptor by single particle electron cryo-microscopy, AMTB-bound state | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoyloxy)methyl]ethyl octadecanoate, CHOLESTEROL HEMISUCCINATE, N-(3-aminopropyl)-2-[(3-methylphenyl)methoxy]-N-[(thiophen-2-yl)methyl]benzamide, ... | | Authors: | Diver, M.M, Cheng, Y, Julius, D. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into TRPM8 inhibition and desensitization.
Science, 365, 2019
|
|
5FAJ
 
 | |
6I08
 
 | | THE GLIC PENTAMERIC LIGAND-GATED ION CHANNEL MUTANT E243C-I201W | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2018-10-25 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Electrostatics, proton sensor, and networks governing the gating transition in GLIC, a proton-gated pentameric ion channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5MUR
 
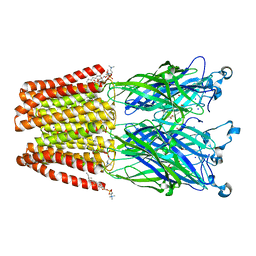 | | X-ray structure of the F14'A mutant of GLIC in complex with propofol | | Descriptor: | 2,6-BIS(1-METHYLETHYL)PHENOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Sauguet, L, Fourati, Z, Delarue, M. | | Deposit date: | 2017-01-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for a Bimodal Allosteric Mechanism of General Anesthetic Modulation in Pentameric Ligand-Gated Ion Channels.
Cell Rep, 23, 2018
|
|
6F15
 
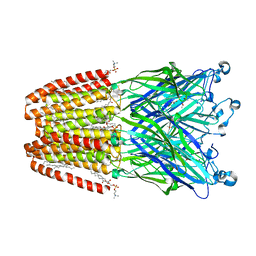 | | GLIC mutant H127Q | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Full mutational mapping of titratable residues helps to identify proton-sensors involved in the control of channel gating in the Gloeobacter violaceus pentameric ligand-gated ion channel.
PLoS Biol., 15, 2017
|
|
3OD9
 
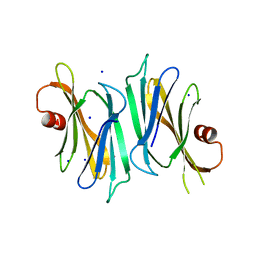 | | Crystal structure of PliI-Ah, periplasmic lysozyme inhibitor of I-type lysozyme from Aeromonas hydrophyla | | Descriptor: | POTASSIUM ION, Putative exported protein, SODIUM ION | | Authors: | Leysen, S, Van Herreweghe, J.M, Callewaert, L, Heirbaut, M, Buntinx, P, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2010-08-11 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.411 Å) | | Cite: | Molecular Basis of Bacterial Defense against Host Lysozymes: X-ray Structures of Periplasmic Lysozyme Inhibitors PliI and PliC.
J.Mol.Biol., 405, 2011
|
|
4LCZ
 
 | | Crystal structure of a multilayer-packed major light-harvesting complex | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wan, T, Li, M, Chang, W.R. | | Deposit date: | 2013-06-24 | | Release date: | 2014-05-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a multilayer packed major light-harvesting complex: implications for grana stacking in higher plants.
Mol Plant, 7, 2014
|
|
6F16
 
 | | GLIC mutant H277Q | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Full mutational mapping of titratable residues helps to identify proton-sensors involved in the control of channel gating in the Gloeobacter violaceus pentameric ligand-gated ion channel.
PLoS Biol., 15, 2017
|
|
