1Z2J
 
 | |
5VB9
 
 | | IL-17A in complex with peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Interleukin-17A, ... | | Authors: | Antonysamy, S, Russell, M, Zhang, A, Groshong, C, Manglicmot, D, Lu, F, Benach, J, Wasserman, S.R, Zhang, F, Afshar, S, Bina, H, Broughton, H, Chalmers, M, Dodge, J, Espada, A, Jones, S, Ting, J.P, Woodman, M. | | Deposit date: | 2017-03-28 | | Release date: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Utilization of peptide phage display to investigate hotspots on IL-17A and what it means for drug discovery.
PLoS ONE, 13, 2018
|
|
3FEL
 
 | |
1ZKG
 
 | |
5UR2
 
 | |
5URY
 
 | | Crystal structure of Frizzled 5 CRD in complex with PAM | | Descriptor: | Frizzled-5, PALMITOLEIC ACID, alpha-L-fucopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mukund, S, Nile, A.H, Stanger, K, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3C3C
 
 | | Crystal Structure of human phosphoglycerate kinase bound to 3-phosphoglycerate and L-CDP | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Arold, S.T, Gondeau, C, Lionne, C, Chaloin, L. | | Deposit date: | 2008-01-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the lack of enantioselectivity of human 3-phosphoglycerate kinase
Nucleic Acids Res., 36, 2008
|
|
5V6R
 
 | | Structure of Plexin D1 intracellular domain | | Descriptor: | Plexin-D1 | | Authors: | Shang, G, Zhang, X. | | Deposit date: | 2017-03-17 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure analyses reveal a regulated oligomerization mechanism of the PlexinD1/GIPC/myosin VI complex.
Elife, 6, 2017
|
|
3C3X
 
 | |
3CAW
 
 | | Crystal structure of o-succinylbenzoate synthase from Bdellovibrio bacteriovorus liganded with Mg | | Descriptor: | MAGNESIUM ION, o-succinylbenzoate synthase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1K3Q
 
 | | NMR structure of the FHA1 Domain of Rad53 in Complex with a Rad9-derived Phosphothreonine (at T192) Peptide | | Descriptor: | DNA repair protein Rad9, Protein Kinase SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
5DW7
 
 | | Crystal structure of the unliganded geosmin synthase N-terminal domain from Streptomyces coelicolor | | Descriptor: | Germacradienol/geosmin synthase | | Authors: | Lombardi, P.M, Harris, G.G, Pemberton, T.A, Matsui, T, Weiss, T.M, Cole, K.E, Koksal, M, Murphy, F.V, Vedula, L.S, Chou, W.K, Cane, D.E, Christianson, D.W. | | Deposit date: | 2015-09-22 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural Studies of Geosmin Synthase, a Bifunctional Sesquiterpene Synthase with alpha alpha Domain Architecture That Catalyzes a Unique Cyclization-Fragmentation Reaction Sequence.
Biochemistry, 54, 2015
|
|
1JM4
 
 | | NMR Structure of P/CAF Bromodomain in Complex with HIV-1 Tat Peptide | | Descriptor: | HIV-1 Tat Peptide, P300/CBP-associated Factor | | Authors: | Mujtaba, S, He, Y, Zeng, L, Farooq, A, Carlson, J.E, Ott, M, Verdin, E, Zhou, M.-M. | | Deposit date: | 2001-07-17 | | Release date: | 2002-07-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain
Mol.Cell, 9, 2002
|
|
1N4O
 
 | |
5SWA
 
 | |
3FKN
 
 | | P38 kinase crystal structure in complex with RO7125 | | Descriptor: | 3-[2-chloro-5-(methylsulfonyl)phenyl]-6-(2,4-difluorophenoxy)-1H-pyrazolo[3,4-d]pyrimidine, Mitogen-activated protein kinase 14 | | Authors: | Kuglstatter, A, Ghate, M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping Binding Pocket Volume: Potential Applications towards Ligand Design and Selectivity
To be Published
|
|
2AEO
 
 | | Crystal structure of cisplatinated bovine Cu,Zn superoxide dismutase | | Descriptor: | COPPER (II) ION, Cisplatin, Superoxide dismutase [Cu-Zn], ... | | Authors: | Calderone, V, Casini, A, Mangani, S, Messori, L, Orioli, P.L. | | Deposit date: | 2005-07-23 | | Release date: | 2006-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural investigation of cisplatin-protein interactions: selective platination of His19 in a cuprozinc superoxide dismutase.
Angew. Chem. Int. Ed. Engl., 45, 2006
|
|
3FFV
 
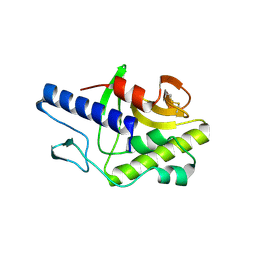 | | Crystal Structure Analysis of Syd | | Descriptor: | Protein syd | | Authors: | Maurus, R, Brayer, G.D. | | Deposit date: | 2008-12-04 | | Release date: | 2009-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, Binding, and Activity of Syd, a SecY-interacting Protein
J.Biol.Chem., 284, 2009
|
|
2A6A
 
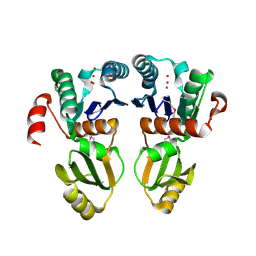 | |
2A6Y
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), tetragonal crystal form | | Descriptor: | Emp47p (form1), SULFATE ION | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6V
 
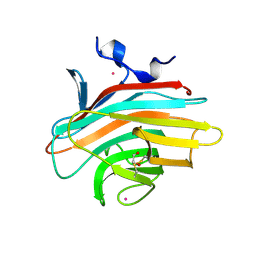 | | Crystal structure of Emp46p carbohydrate recognition domain (CRD), potassium-bound form | | Descriptor: | 1,2-ETHANEDIOL, Emp46p, POTASSIUM ION | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
5W2V
 
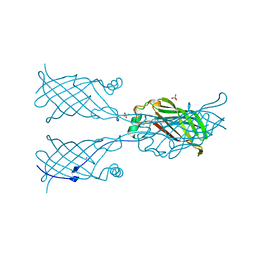 | |
3AUR
 
 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with Yne-diene type analog of active 14-epi-2beta-methyl-19-norvitamin D3 | | Descriptor: | (1R,2S,3R)-5-[2-[(1R,3aS,7aR)-1-[(2R)-6-hydroxy-6-methyl-heptan-2-yl]-7a-methyl-1,2,3,3a,6,7-hexahydroinden-4-yl]ethynyl]-2-methyl-cyclohex-4-ene-1,3-diol, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2011-02-15 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Development of 14-epi-19-nortachysterol and its unprecedented binding configuration for the human vitamin D receptor
J.Am.Chem.Soc., 133, 2011
|
|
2DTU
 
 | | Crystal structure of the beta hairpin loop deletion variant of RB69 gp43 in complex with DNA containing an abasic site analog | | Descriptor: | 5'-D(*CP*GP*(3DR)P*CP*TP*TP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*CP*G)-3', 5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*AP*AP*GP*A)-3', DNA polymerase | | Authors: | Aller, P, Hogg, M, Konigsberg, W, Wallace, S.S, Doublie, S. | | Deposit date: | 2006-07-15 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural and biochemical investigation of the role in proofreading of a beta hairpin loop found in the exonuclease domain of a replicative DNA polymerase of the B family.
J.Biol.Chem., 282, 2007
|
|
1J4O
 
 | | REFINED NMR STRUCTURE OF THE FHA1 DOMAIN OF YEAST RAD53 | | Descriptor: | PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
