1WIZ
 
 | | Solution structure of the first CUT domain of KIAA1034 protein | | Descriptor: | DNA-binding protein SATB2 | | Authors: | Inoue, K, Nameki, S, Hayashi, F, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first CUT domain of KIAA1034 protein
To be Published
|
|
2Z3Y
 
 | | Crystal structure of Lysine-specific demethylase1 | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL (2R,3S,4S)-5-[7,8-DIMETHYL-2,4-DIOXO-5-(3-PHENYLPROPANOYL)-1,3,4,5-TETRAHYDROBENZO[G]PTERIDIN-10(2H)-YL]-2,3,4-TRIHYDROXYPENTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Mimasu, S, Sengoku, T, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-06-08 | | Release date: | 2008-01-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of histone demethylase LSD1 and tranylcypromine at 2.25A
Biochem.Biophys.Res.Commun., 366, 2008
|
|
1WM2
 
 | |
3O2T
 
 | |
1EWD
 
 | | FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE FROM RABBIT MUSCLE | | Descriptor: | FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE | | Authors: | Maurady, A, Sygusch, J. | | Deposit date: | 2000-04-25 | | Release date: | 2001-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.464 Å) | | Cite: | A conserved glutamate residue exhibits multifunctional catalytic roles in
D-fructose-1,6-bisphosphate aldolases.
J.Biol.Chem., 277, 2002
|
|
1WTB
 
 | | Complex structure of the C-terminal RNA-binding domain of hnRNP D (AUF1) with telomere DNA | | Descriptor: | 5'-D(P*TP*AP*GP*G)-3', Heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Enokizono, Y, Konishi, Y, Nagata, K, Ouhashi, K, Uesugi, S, Ishikawa, F, Katahira, M. | | Deposit date: | 2004-11-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D
J.Biol.Chem., 280, 2005
|
|
3NNC
 
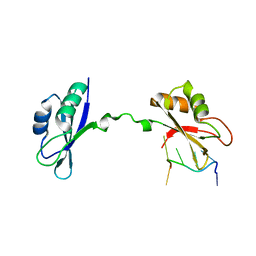 | | Crystal Structure of CUGBP1 RRM1/2-RNA Complex | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*UP*GP*UP*GP*UP*GP*UP*UP*GP*UP*GP*UP*G)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2005 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
1EX5
 
 | | FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE FROM RABBIT MUSCLE | | Descriptor: | FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE | | Authors: | Maurady, A, Sygusch, J. | | Deposit date: | 2000-04-25 | | Release date: | 2001-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A conserved glutamate residue exhibits multifunctional catalytic roles in
D-fructose-1,6-bisphosphate aldolases.
J.Biol.Chem., 277, 2002
|
|
1SAV
 
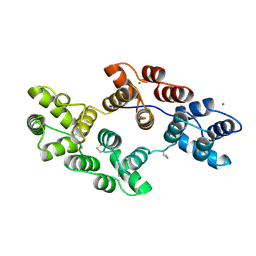 | | HUMAN ANNEXIN V WITH PROLINE SUBSTITUTION BY THIOPROLINE | | Descriptor: | ANNEXIN V, CALCIUM ION | | Authors: | Medrano, F.J, Minks, C, Budisa, N, Huber, R. | | Deposit date: | 1997-11-24 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal and molecular structure of human annexin V after refinement. Implications for structure, membrane binding and ion channel formation of the annexin family of proteins.
J.Mol.Biol., 223, 1992
|
|
1WHW
 
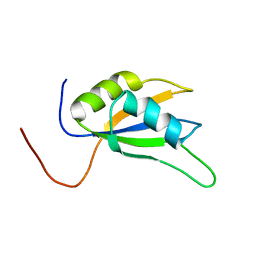 | | Solution structure of the N-terminal RNA binding domain from hypothetical protein BAB23448 | | Descriptor: | hypothetical protein RIKEN CDNA 1200009A02 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal RNA binding domain from hypothetical protein BAB23448
To be Published
|
|
3O50
 
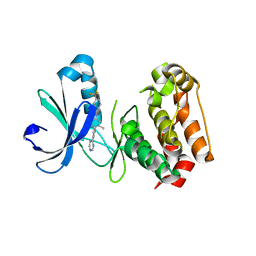 | | Crystal structure of benzamide 9 bound to AuroraA | | Descriptor: | N-{3-methyl-4-[(3-pyrimidin-4-ylpyridin-2-yl)oxy]phenyl}-3-(trifluoromethyl)benzamide, cDNA FLJ58295, highly similar to Serine/threonine-protein kinase 6 | | Authors: | Huang, X. | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a potent, selective, and orally bioavailable pyridinyl-pyrimidine phthalazine aurora kinase inhibitor.
J.Med.Chem., 53, 2010
|
|
1F3C
 
 | |
1X0F
 
 | | Complex structure of the C-terminal RNA-binding domain of hnRNP D(AUF1) with telomeric DNA | | Descriptor: | 5'-D(P*TP*AP*GP*G)-3', Heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Enokizono, Y, Konishi, Y, Nagata, K, Ouhashi, K, Uesugi, S, Ishikawa, F, Katahira, M. | | Deposit date: | 2005-03-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D.
J.Biol.Chem., 280, 2005
|
|
3A6P
 
 | | Crystal structure of Exportin-5:RanGTP:pre-miRNA complex | | Descriptor: | 13-mer peptide, Exportin-5, GTP-binding nuclear protein Ran, ... | | Authors: | Okada, C, Yamashita, E, Lee, S.J, Shibata, S, Katahira, J, Nakagawa, A, Yoneda, Y, Tsukihara, T. | | Deposit date: | 2009-09-07 | | Release date: | 2009-12-08 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | A high-resolution structure of the pre-microRNA nuclear export machinery
Science, 326, 2009
|
|
1X4A
 
 | | Solution structure of RRM domain in splicing factor SF2 | | Descriptor: | splicing factor, arginine/serine-rich 1 (splicing factor 2, alternate splicing factor) variant | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in splicing factor SF2
To be Published
|
|
3NRM
 
 | | Imidazo[1,2-a]pyrazine-based Aurora Kinase Inhibitors | | Descriptor: | N-(3-methylisothiazol-5-yl)-3-(1H-pyrazol-4-yl)imidazo[1,2-a]pyrazin-8-amine, Serine/threonine-protein kinase 6 | | Authors: | Hruza, A. | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery of imidazo[1,2-a]pyrazine-based Aurora kinase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3NNA
 
 | | Crystal Structure of CUGBP1 RRM1/2-RNA Complex | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*GP*UP*UP*GP*UP*UP*UP*UP*GP*UP*UP*U)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
1U35
 
 | | Crystal structure of the nucleosome core particle containing the histone domain of macroH2A | | Descriptor: | H2A histone family, Hist1h4i protein, Histone H3.1, ... | | Authors: | Chakravarthy, S, Gundimella, S.K, Caron, C, Perche, P.Y, Pehrson, J.R, Khochbin, S, Luger, K. | | Deposit date: | 2004-07-20 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural characterization of the histone variant macroH2A.
Mol.Cell.Biol., 25, 2005
|
|
1DZ1
 
 | | Mouse HP1 (M31) C terminal (shadow chromo) domain | | Descriptor: | MODIFIER 1 PROTEIN | | Authors: | Brasher, S.V, Smith, B.O, Fogh, R.H, Nietlispach, D, Thiru, A, Nielsen, P.R, Broadhurst, R.W, Ball, L.J, Murzina, N, Laue, E.D. | | Deposit date: | 2000-02-11 | | Release date: | 2000-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of Mouse Hp1 Suggests a Unique Mode of Single Peptide Recognition by the Shadow Chromo Domain Dimer
Embo J., 19, 2000
|
|
1U41
 
 | | Crystal structure of YLGV mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
5TA8
 
 | | Crystal structure of PLK1 in complex with a novel 5,6-dihydroimidazolo[1,5-f]pteridine inhibitor | | Descriptor: | 4-[(9-cyclopentyl-7,7-difluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Serine/threonine-protein kinase PLK1, ZINC ION | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design and SAR development of 5,6-dihydroimidazolo[1,5-f]pteridine derivatives as novel Polo-like kinase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5TC4
 
 | | Crystal structure of human mitochondrial methylenetetrahydrofolate dehydrogenase-cyclohydrolase (MTHFD2) in complex with LY345899 and cofactors | | Descriptor: | 4-(7-AMINO-9-HYDROXY-1-OXO-3,3A,4,5-TETRAHYDRO-2,5,6,8,9B-PENTAAZA-CYCLOPENTA[A]NAPHTHALEN-2-YL)-PHENYLCARBONYL-GLUTAMI C ACID, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | Authors: | Gustafsson, R, Jemth, A.-S, Gustafsson Sheppard, N, Farnegardh, K, Loseva, O, Wiita, E, Bonagas, N, Dahllund, L, Llona-Minguez, S, Haggblad, M, Henriksson, M, Andersson, Y, Homan, E, Helleday, T, Stenmark, P. | | Deposit date: | 2016-09-14 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of the Emerging Cancer Target MTHFD2 in Complex with a Substrate-Based Inhibitor.
Cancer Res., 77, 2017
|
|
5TLZ
 
 | | Fructose-1,6-bisphosphate aldolase from rabbit muscle in complex with the inhibitor naphthalene 2,6-bisphosphate | | Descriptor: | Fructose-bisphosphate aldolase A, GLYCEROL, naphthalene-2,6-diyl bis[dihydrogen (phosphate)] | | Authors: | Heron, P.W, Sygusch, J. | | Deposit date: | 2016-10-12 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Bisphosphonate Inhibitors of Mammalian Glycolytic Aldolase.
J.Med.Chem., 61, 2018
|
|
3ACA
 
 | |
1FT3
 
 | | CRYSTAL STRUCTURE OF TRUNCATED RHOGDI K141A MUTANT | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1 | | Authors: | Longenecker, K.L, Garrard, S.M, Sheffield, P.J, Derewenda, Z.S. | | Deposit date: | 2000-09-11 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystallization by rational mutagenesis of surface residues: Lys to Ala mutations promote crystallization of RhoGDI.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
