2YI7
 
 | | Structural characterization of 5-Aryl-4-(5-substituted-2-4- dihydroxyphenyl)-1,2,3-thiadiazole Hsp90 inhibitors. | | Descriptor: | 4-CHLORO-6-[5-(4-ETHOXYPHENYL)-1,2,3-THIADIAZOL-4-YL BENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP 90-ALPHA, MAGNESIUM ION | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2011-05-10 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Co-Crystalization and in Vitro Biological Characterization of 5-Aryl-4-(5-Substituted-2-4-Dihydroxyphenyl)-1,2,3-Thiadiazole Hsp90 Inhibitors.
Plos One, 7, 2012
|
|
1XMZ
 
 | | Crystal structure of the dark state of kindling fluorescent protein kfp from anemonia sulcata | | Descriptor: | BETA-MERCAPTOETHANOL, GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Quillin, M.L, Anstrom, D.M, Shu, X, O'Leary, S, Kallio, K, Chudakov, D.M, Remington, S.J. | | Deposit date: | 2004-10-04 | | Release date: | 2005-04-19 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Kindling Fluorescent Protein from Anemonia sulcata: Dark-State Structure at 1.38 Resolution
Biochemistry, 44, 2005
|
|
1RNK
 
 | |
3TQW
 
 | | Structure of a ABC transporter, periplasmic substrate-binding protein from Coxiella burnetii | | Descriptor: | METHIONINE, Methionine-binding protein, SULFATE ION | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TRC
 
 | | Structure of the GAF domain from a phosphoenolpyruvate-protein phosphotransferase (ptsP) from Coxiella burnetii | | Descriptor: | PHOSPHATE ION, Phosphoenolpyruvate-protein phosphotransferase, SODIUM ION | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
4CAU
 
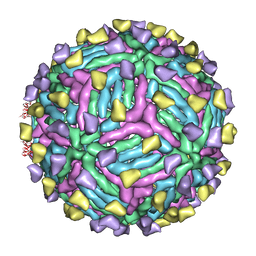 | | THREE-DIMENSIONAL STRUCTURE OF DENGUE VIRUS SEROTYPE 1 COMPLEXED WITH 2 HMAB 14C10 FAB | | Descriptor: | ENVELOPE PROTEIN E, FAB 14C10 | | Authors: | Teoh, E.P, Kukkaro, P, Teo, E.W, Lim, A.P, Tan, T.T, Yip, A, Schul, W, Aung, M, Kostyuchenko, V.A, Leo, Y.S, Chan, S.H, Smith, K.G, Chan, A.H, Zou, G, Ooi, E.E, Kemeny, D.M, Tan, G.K, Ng, J.K, Ng, M.L, Alonso, S, Fisher, D, Shi, P.Y, Hanson, B.J, Lok, S.M, Macary, P.A. | | Deposit date: | 2013-10-09 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | The Structural Basis for Serotype-Specific Neutralization of Dengue Virus by a Human Antibody.
Sci.Trans.Med, 4, 2012
|
|
4C2P
 
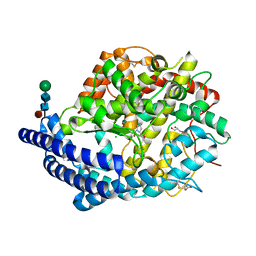 | | Crystal structure of human testis angiotensin-I converting enzyme mutant R522K in complex with captopril | | Descriptor: | ACETATE ION, ANGIOTENSIN-CONVERTING ENZYME, CHLORIDE ION, ... | | Authors: | Masuyer, G, Yates, C.J, Schwager, S.L.U, Mohd, A, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2013-08-19 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular and Thermodynamic Mechanisms of the Chloride Dependent Human Angiotensin-I Converting Enzyme (Ace)
J.Biol.Chem., 289, 2014
|
|
3TQH
 
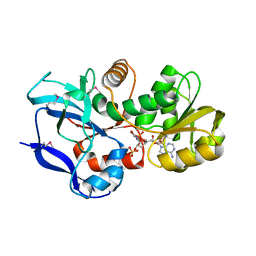 | | Structure of the quinone oxidoreductase from Coxiella burnetii | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Quinone oxidoreductase, SULFATE ION | | Authors: | Franklin, M.C, Cheung, J, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TQP
 
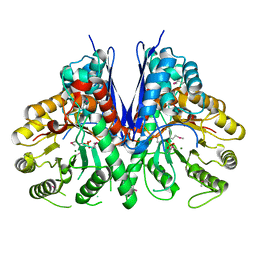 | | Structure of an enolase (eno) from Coxiella burnetii | | Descriptor: | Enolase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Rudolph, M, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TRI
 
 | | Structure of a pyrroline-5-carboxylate reductase (proC) from Coxiella burnetii | | Descriptor: | CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PHOSPHATE ION, ... | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
4C2Q
 
 | | Crystal structure of human testis angiotensin-I converting enzyme mutant R522K | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ACETATE ION, ANGIOTENSIN-CONVERTING ENZYME, ... | | Authors: | Masuyer, G, Yates, C.J, Schwager, S.L.U, Mohd, A, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2013-08-19 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular and Thermodynamic Mechanisms of the Chloride Dependent Human Angiotensin-I Converting Enzyme (Ace)
J.Biol.Chem., 289, 2014
|
|
3TR0
 
 | | Structure of guanylate kinase (gmk) from Coxiella burnetii | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Guanylate kinase, SULFATE ION | | Authors: | Cheung, J, Franklin, M, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TRE
 
 | | Structure of a translation elongation factor P (efp) from Coxiella burnetii | | Descriptor: | Elongation factor P | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3T0X
 
 | | Fluorogen Activating Protein M8VLA4(S55P) in complex with dimethylindole red | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-sulfopropyl)-4-[(1E,3E)-3-(1,3,3-trimethyl-1,3-dihydro-2H-indol-2-ylidene)prop-1-en-1-yl]quinolinium, Immunoglobulin variable lambda domain M8VLA4(S55P), ... | | Authors: | Stanfield, R, Senutovitch, N, Bhattacharyya, S, Rule, G, Wilson, I.A, Armitage, B, Waggoner, A.S, Berget, P. | | Deposit date: | 2011-07-20 | | Release date: | 2012-03-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A variable light domain fluorogen activating protein homodimerizes to activate dimethylindole red.
Biochemistry, 51, 2012
|
|
3T0V
 
 | | Unliganded fluorogen activating protein M8VL | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Stanfield, R, Senutovitch, N, Bhattacharyya, S, Rule, G, Wilson, I.A, Armitage, B, Waggoner, A.S, Berget, P. | | Deposit date: | 2011-07-20 | | Release date: | 2012-03-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | A variable light domain fluorogen activating protein homodimerizes to activate dimethylindole red.
Biochemistry, 51, 2012
|
|
1EXD
 
 | | CRYSTAL STRUCTURE OF A TIGHT-BINDING GLUTAMINE TRNA BOUND TO GLUTAMINE AMINOACYL TRNA SYNTHETASE | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLUTAMINE TRNA APTAMER, GLUTAMINYL-TRNA SYNTHETASE, ... | | Authors: | Bullock, T.L, Sherlin, L.D, Perona, J.J. | | Deposit date: | 2000-05-02 | | Release date: | 2000-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tertiary core rearrangements in a tight binding transfer RNA aptamer.
Nat.Struct.Biol., 7, 2000
|
|
2MZV
 
 | |
2YI5
 
 | | Structural characterization of 5-Aryl-4-(5-substituted-2-4- dihydroxyphenyl)-1,2,3-thiadiazole Hsp90 inhibitors. | | Descriptor: | 4-CHLORO-6-[5-(3,4-DIMETHOXYPHENYL)-1,2,3-THIADIAZOL-4-YL]BENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2011-05-10 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Co-Crystalization and in Vitro Biological Characterization of 5-Aryl-4-(5-Substituted-2-4-Dihydroxyphenyl)-1,2,3-Thiadiazole Hsp90 Inhibitors.
Plos One, 7, 2012
|
|
4Q37
 
 | |
1T31
 
 | | A Dual Inhibitor of the Leukocyte Proteases Cathepsin G and Chymase with Therapeutic Efficacy in Animals Models of Inflammation | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[3-({METHYL[1-(2-NAPHTHOYL)PIPERIDIN-4-YL]AMINO}CARBONYL)-2-NAPHTHYL]-1-(1-NAPHTHYL)-2-OXOETHYLPHOSPHONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | de Garavilla, L, Greco, M.N, Giardino, E.C, Wells, G.I, Haertlein, B.J, Kauffman, J.A, Corcoran, T.W, Derian, C.K, Eckardt, A.J, Abraham, W.M, Sukumar, N, Chen, Z, Pineda, A.O, Mathews, F.S, Di Cera, E, Andrade-Gordon, P, Damiano, B.P, Maryanoff, B.E, Pereira, P.J.B, Wang, Z.M, Rubin, H, Huber, R, Bode, W, Schechter, N.M, Strobl, S. | | Deposit date: | 2004-04-23 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel, potent dual inhibitor of the leukocyte proteases cathepsin G and chymase: molecular mechanisms and anti-inflammatory activity in vivo.
J.Biol.Chem., 280, 2005
|
|
4CW5
 
 | |
3W3S
 
 | | Crystal structure of A. aeolicus tRNASec in complex with M. kandleri SerRS | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, PLATINUM (II) ION, SULFATE ION, ... | | Authors: | Itoh, Y, Sekine, S, Yokoyama, S. | | Deposit date: | 2012-12-27 | | Release date: | 2013-02-13 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (3.095 Å) | | Cite: | Tertiary structure of bacterial selenocysteine tRNA
Nucleic Acids Res., 41, 2013
|
|
3TY2
 
 | | Structure of a 5'-nucleotidase (surE) from Coxiella burnetii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-nucleotidase surE | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-23 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.885 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TQX
 
 | | Structure of the 2-amino-3-ketobutyrate coenzyme A ligase (kbl) from Coxiella burnetii | | Descriptor: | 2-amino-3-ketobutyrate coenzyme A ligase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TRD
 
 | | Structure of an alpha-beta serine hydrolase homologue from Coxiella burnetii | | Descriptor: | ACETATE ION, Alpha/beta hydrolase, PHOSPHATE ION, ... | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
