6KBA
 
 | | X-ray structure of human PPARalpha ligand binding domain-Wy14643 co-crystals obtained by co-crystallization | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
1BL4
 
 | | FKBP MUTANT F36V COMPLEXED WITH REMODELED SYNTHETIC LIGAND | | Descriptor: | PROTEIN (FK506 BINDING PROTEIN), {3-[3-(3,4-DIMETHOXY-PHENYL)-1-(1-{1-[2-(3,4,5-TRIMETHOXY-PHENYL)-BUTYRYL]-PIPERIDIN-2YL}-VINYLOXY)-PROPYL]-PHENOXY}-ACETIC ACID | | Authors: | Hatada, M.H, Clackson, T, Yang, W, Rozamus, L.W, Amara, J, Rollins, C.T, Stevenson, L.F, Magari, S.R, Wood, S.A, Courage, N.L, Lu, X, Cerasoli Junior, F, Gilman, M, Holt, D. | | Deposit date: | 1998-07-23 | | Release date: | 1998-09-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6KAY
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW7647 co-crystals obtained by soaking | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB3
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW7647 co-crystals obtained by delipidation and cross-seeding | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KAX
 
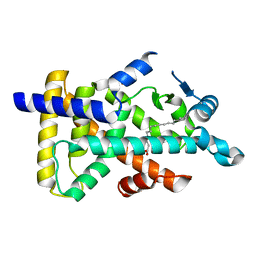 | | X-ray structure of human PPARalpha ligand binding domain-intrinsic fatty acid (E. coli origin) co-crystals obtained by cross-seeding | | Descriptor: | GLYCEROL, PALMITIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB4
 
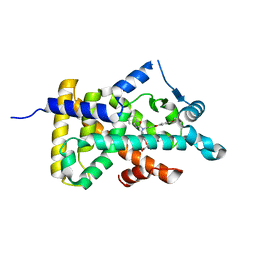 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate co-crystals obtained by delipidation and cross-seeding | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB6
 
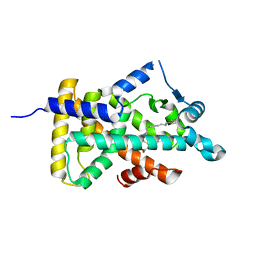 | | X-ray structure of human PPARalpha ligand binding domain-tetradecylthioacetic acid (TTA) co-crystals obtained by delipidation and cross-seeding | | Descriptor: | 2-tetradecylsulfanylethanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.431 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB8
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW7647 co-crystals obtained by cross-seeding | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB7
 
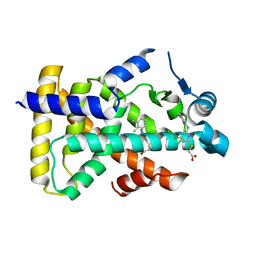 | | X-ray structure of human PPARalpha ligand binding domain-Wy14643 co-crystals obtained by delipidation and cross-seeding | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
4W51
 
 | | T4 Lysozyme L99A with No Ligand Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W9F
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 5) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(4-methyl-1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | van Molle, I, Hewitt, S, Galdeano, C, Gadd, M.S, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
3K6U
 
 | | M. acetivorans Molybdate-Binding Protein (ModA) in Unliganded Open Form | | Descriptor: | Solute-binding protein MA_0280 | | Authors: | Chan, S, Giuroiu, I, Chernishof, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
6CDL
 
 | | HIV-1 wild type protease with GRL-03214A, 6-5-5-ring fused umbrella-like tetrahydropyranofuran as the P2-ligand, a cyclopropylaminobenzothiazole as the P2'-ligand and 3,5-difluorophenylmethyl as the P1-ligand | | Descriptor: | (2aR,4S,4aR,7aR,7bR)-octahydro-2H-1,7-dioxacyclopenta[cd]inden-4-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design and Synthesis of Highly Potent HIV-1 Protease Inhibitors Containing Tricyclic Fused Ring Systems as Novel P2 Ligands: Structure-Activity Studies, Biological and X-ray Structural Analysis.
J. Med. Chem., 61, 2018
|
|
2ZVT
 
 | | Cys285Ser mutant PPARgamma ligand-binding domain complexed with 15-deoxy-delta12,14-prostaglandin J2 | | Descriptor: | (5E,14E)-11-oxoprosta-5,9,12,14-tetraen-1-oic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Waku, T, Oyama, T, Shiraki, T, Morikawa, K. | | Deposit date: | 2008-11-19 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Atomic structure of mutant PPARgamma LBD complexed with 15d-PGJ2: novel modulation mechanism of PPARgamma/RXRalpha function by covalently bound ligands
Febs Lett., 583, 2009
|
|
7ONI
 
 | | Structure of Neddylated CUL5 C-terminal region-RBX2-ARIH2* | | Descriptor: | Cullin-5, E3 ubiquitin-protein ligase ARIH2, NEDD8, ... | | Authors: | Kostrhon, S.P, prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CUL5-ARIH2 E3-E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation.
Nat.Chem.Biol., 17, 2021
|
|
1AEU
 
 | | SPECIFICITY OF LIGAND BINDING IN A POLAR CAVITY OF CYTOCHROME C PEROXIDASE (2-METHYLIMIDAZOLE) | | Descriptor: | 2-METHYLIMIDAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-25 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A ligand-gated, hinged loop rearrangement opens a channel to a buried artificial protein cavity.
Nat.Struct.Biol., 3, 1996
|
|
1AA4
 
 | | SPECIFICITY OF LIGAND BINDING IN A BURIED POLAR CAVITY OF CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-01-22 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A ligand-gated, hinged loop rearrangement opens a channel to a buried artificial protein cavity.
Nat.Struct.Biol., 3, 1996
|
|
1AES
 
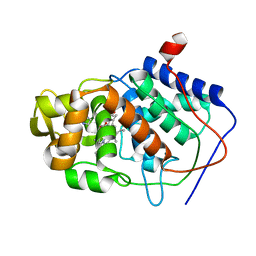 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (IMIDAZOLE) | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-25 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A ligand-gated, hinged loop rearrangement opens a channel to a buried artificial protein cavity.
Nat.Struct.Biol., 3, 1996
|
|
6CDJ
 
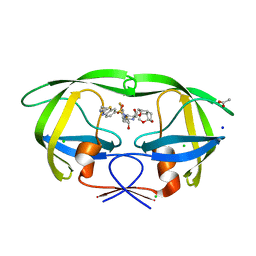 | | HIV-1 wild type protease with GRL-03314A, 6-5-5-ring fused umbrella-like tetrahydropyranofuran as the P2-ligand, a cyclopropylaminobenzothiazole as the P2'-ligand and 3,5-difluorophenylmethyl as the P1-ligand | | Descriptor: | (2aS,4R,4aS,7aS,7bS)-octahydro-2H-1,7-dioxacyclopenta[cd]inden-4-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Design and Synthesis of Highly Potent HIV-1 Protease Inhibitors Containing Tricyclic Fused Ring Systems as Novel P2 Ligands: Structure-Activity Studies, Biological and X-ray Structural Analysis.
J. Med. Chem., 61, 2018
|
|
4P9V
 
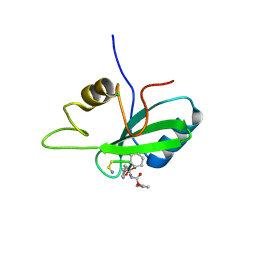 | | Grb2 SH2 complexed with a pTyr-Ac6cN-Asn tripeptide | | Descriptor: | CHLORIDE ION, Growth factor receptor-bound protein 2, PHQ-PTR-02K-ASN-NH2 | | Authors: | Clements, J.H, Martin, S.F. | | Deposit date: | 2014-04-06 | | Release date: | 2014-06-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Protein-ligand interactions: Probing the energetics of a putative cation-pi interaction.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4P9Z
 
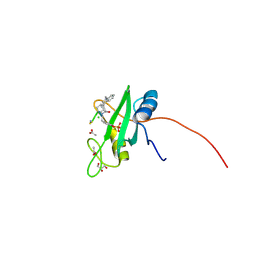 | |
8WQF
 
 | | cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CUX1 (conformation 2) | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQE
 
 | | Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CUX1 (conformation 1) | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQC
 
 | | cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CDK5R1 | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQG
 
 | | cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 1) | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
