6DRB
 
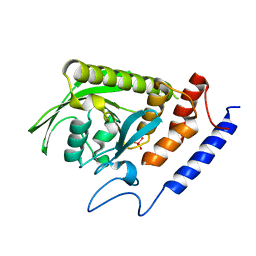 | | Crystal Structure of the YopH PTP1B WPD loop Chimera 3 PTPase bound to tungstate | | Descriptor: | TUNGSTATE(VI)ION, Targeted effector protein | | Authors: | Morales, Y, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.745 Å) | | Cite: | A YopH PTP1B Chimera Shows the Importance of the WPD-Loop Sequence to the Activity, Structure, and Dynamics of Protein Tyrosine Phosphatases.
Biochemistry, 57, 2018
|
|
6DL1
 
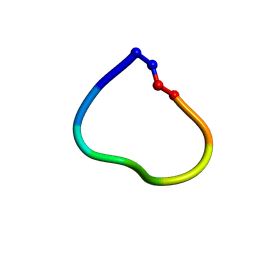 | | Racemic structure of jatrophidin, an orbitide from Jatropha curcas | | Descriptor: | jatrophidin | | Authors: | Wang, C.K, King, G.J, Ramalho, S.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-11-14 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.029 Å) | | Cite: | Synthesis, Racemic X-ray Crystallographic, and Permeability Studies of Bioactive Orbitides from Jatropha Species.
J. Nat. Prod., 81, 2018
|
|
6DO6
 
 | | NMR solution structure of wild type apo hFABP1 at 308 K | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-09 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
6DBU
 
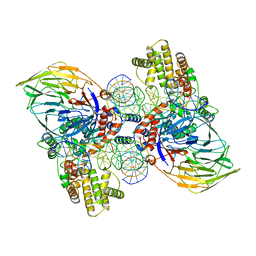 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBI
 
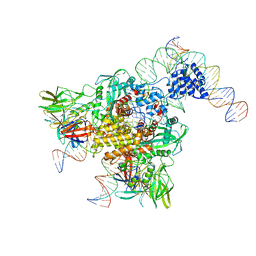 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS nicked DNA intermediates | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS signal end, Forward strand of 23-RSS signal end, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DFR
 
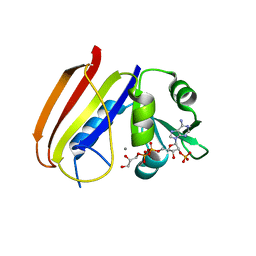 | |
6KQQ
 
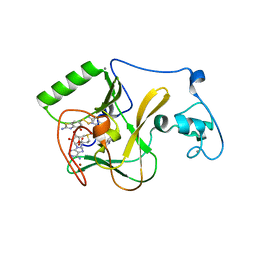 | | NSD1 SET domain in complex with BT3 and SAM | | Descriptor: | 2-azanyl-6-[(2-azanyl-4-oxidanyl-1,3-benzothiazol-6-yl)disulfanyl]-1,3-benzothiazol-4-ol, 2-azanyl-6-sulfanyl-1,3-benzothiazol-4-ol, CALCIUM ION, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2019-08-18 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent inhibition of NSD1 histone methyltransferase.
Nat.Chem.Biol., 16, 2020
|
|
6DMX
 
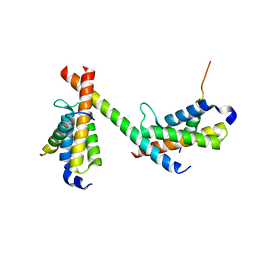 | | HBZ56 in complex with KIX and c-Myb | | Descriptor: | BZIP factor, CREB-binding protein, Transcriptional activator Myb | | Authors: | Yang, K, Wright, P.E, Stanfield, R.L. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cooperative regulation of KIX-mediated transcription pathways by the HTLV-1 HBZ activation domain.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DNQ
 
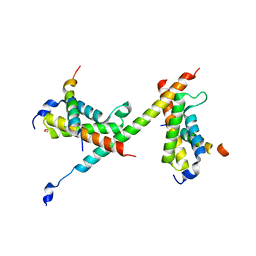 | | HBZ77 in complex with KIX and c-Myb | | Descriptor: | 1,2-ETHANEDIOL, BZIP factor, CREB-binding protein, ... | | Authors: | Yang, K, Wright, P.E, Stanfield, R.L. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for cooperative regulation of KIX-mediated transcription pathways by the HTLV-1 HBZ activation domain.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DL0
 
 | | Crystal structure of pohlianin C, an orbitide from Jatropha pohliana | | Descriptor: | pohlianin C | | Authors: | Wang, C.K, King, G.J, Ramalho, S.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-11-07 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Synthesis, Racemic X-ray Crystallographic, and Permeability Studies of Bioactive Orbitides from Jatropha Species.
J. Nat. Prod., 81, 2018
|
|
6L54
 
 | | Structure of SMG189 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein SMG8, ... | | Authors: | Xu, Y, Qi, Y. | | Deposit date: | 2019-10-22 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Cryo-EM structure of SMG1-SMG8-SMG9 complex.
Cell Res., 29, 2019
|
|
6DR1
 
 | | YopH PTP1B Chimera 2 PTPase | | Descriptor: | Targeted effector protein | | Authors: | Morales, Y, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A YopH PTP1B Chimera Shows the Importance of the WPD-Loop Sequence to the Activity, Structure, and Dynamics of Protein Tyrosine Phosphatases.
Biochemistry, 57, 2018
|
|
6DR7
 
 | | YopH PTP1B WPD loop Chimera 2 PTPase bound to vanadate | | Descriptor: | ACETATE ION, Targeted effector protein, VANADATE ION | | Authors: | Moise, G, Morales, Y, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | A YopH PTP1B Chimera Shows the Importance of the WPD-Loop Sequence to the Activity, Structure, and Dynamics of Protein Tyrosine Phosphatases.
Biochemistry, 57, 2018
|
|
7TPI
 
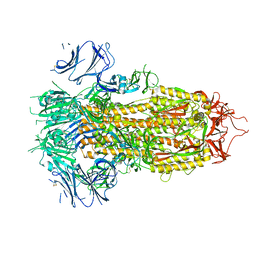 | | SARS-CoV-2 E406W mutant Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Addetia, A, Veesler, D. | | Deposit date: | 2022-01-25 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | SARS-CoV-2 E406W mutant Spike ectodomain
To Be Published
|
|
6DBJ
 
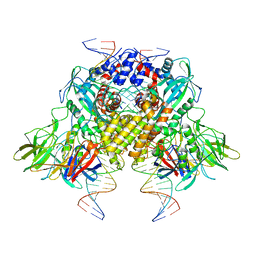 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS nicked DNA intermediates | | Descriptor: | CALCIUM ION, Forward stand of RSS signal end, Forward strand of coding flank, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6L9W
 
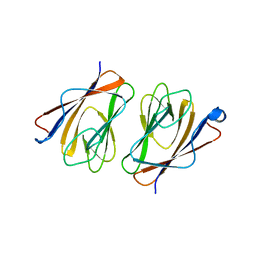 | | Crystal structure of mouse TIFA (T9E/C36S mutant) | | Descriptor: | TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2019-11-11 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of TIFA: Insight into TIFA-dependent signal transduction in innate immunity.
Sci Rep, 10, 2020
|
|
6L49
 
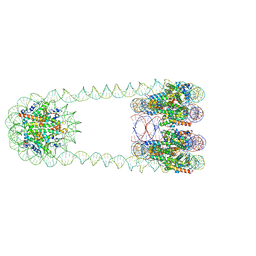 | | H3-CA-H3 tri-nucleosome with the 22 base-pair linker DNA | | Descriptor: | DNA (485-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Takizawa, Y, Ho, C.-H, Tachiwana, H, Matsunami, H, Ohi, M, Wolf, M, Kurumizaka, H. | | Deposit date: | 2019-10-16 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (18.9 Å) | | Cite: | Cryo-EM Structures of Centromeric Tri-nucleosomes Containing a Central CENP-A Nucleosome.
Structure, 28, 2020
|
|
6L5G
 
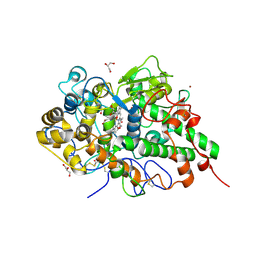 | | Crystal structure of yak lactoperoxidase with disordered heme moiety at 2.50 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, P.K, Rani, C, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Potassium-induced partial inhibition of lactoperoxidase: structure of the complex of lactoperoxidase with potassium ion at 2.20 angstrom resolution.
J.Biol.Inorg.Chem., 26, 2021
|
|
6L9V
 
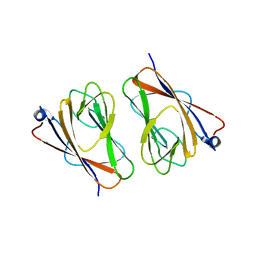 | | Crystal structure of mouse TIFA (T9D/C36S mutant) | | Descriptor: | TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2019-11-11 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural analysis of TIFA: Insight into TIFA-dependent signal transduction in innate immunity.
Sci Rep, 10, 2020
|
|
6LDI
 
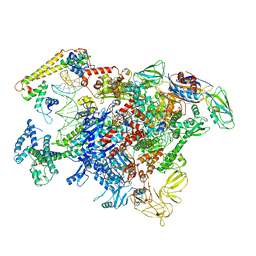 | |
6KU9
 
 | |
6DB7
 
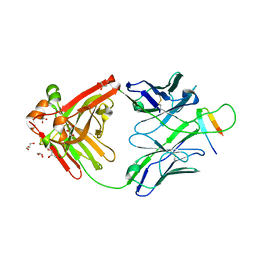 | |
6DBO
 
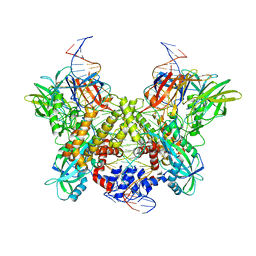 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of substrate RSS DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6KQP
 
 | | NSD1 SET domain in complex with SAM | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific, S-ADENOSYLMETHIONINE, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2019-08-18 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Covalent inhibition of NSD1 histone methyltransferase.
Nat.Chem.Biol., 16, 2020
|
|
6DKZ
 
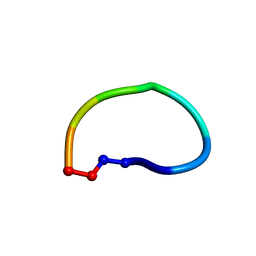 | | Racemic structure of ribifolin, an orbitide from Jatropha ribifolia | | Descriptor: | ribifolin | | Authors: | Wang, C.K, King, G.J, Ramalho, S.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-11-14 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Synthesis, Racemic X-ray Crystallographic, and Permeability Studies of Bioactive Orbitides from Jatropha Species.
J. Nat. Prod., 81, 2018
|
|
