8GHE
 
 | | The structure of h12-LOX in tetrameric form bound to endogenous inhibitor oleoyl-CoA | | Descriptor: | FE (II) ION, Polyunsaturated fatty acid lipoxygenase ALOX12, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name) | | Authors: | Black, K.A, Mobbs, J.I, Venugopal, H, Thal, D.M, Glukhova, A. | | Deposit date: | 2023-03-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | Cryo-EM structures of human arachidonate 12S-lipoxygenase bound to endogenous and exogenous inhibitors.
Blood, 142, 2023
|
|
8GHB
 
 | | The structure of h12-LOX in monomeric form | | Descriptor: | FE (II) ION, Polyunsaturated fatty acid lipoxygenase ALOX12 | | Authors: | Black, K.A, Mobbs, J.I, Venugopal, H, Thal, D.M, Glukhova, A. | | Deposit date: | 2023-03-09 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Cryo-EM structures of human arachidonate 12S-lipoxygenase bound to endogenous and exogenous inhibitors.
Blood, 142, 2023
|
|
1FAD
 
 | | DEATH DOMAIN OF FAS-ASSOCIATED DEATH DOMAIN PROTEIN, RESIDUES 89-183 | | Descriptor: | PROTEIN (FADD PROTEIN) | | Authors: | Jeong, E.-J, Bang, S, Lee, T.H, Park, Y.-I, Sim, W.-S, Kim, K.-S. | | Deposit date: | 1999-03-23 | | Release date: | 1999-07-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of FADD death domain. Structural basis of death domain interactions of Fas and FADD.
J.Biol.Chem., 274, 1999
|
|
8GRJ
 
 | | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yoshida, H, Kojima, K, Tsugawa, W, Okuda-Shimazaki, J, Kerrigan, J.A, Sode, K. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase in complex with gluconolactone
To Be Published
|
|
6VCA
 
 | | TB38 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-nucleotidase, ... | | Authors: | Zhou, Y.F, Lord, D.M. | | Deposit date: | 2019-12-20 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | A highly potent CD73 biparatopic antibody blocks organization of the enzyme active site through dual mechanisms.
J.Biol.Chem., 295, 2020
|
|
3AFF
 
 | | Crystal structure of the HsaA monooxygenase from M. tuberculosis | | Descriptor: | Hydroxylase, putative | | Authors: | D'Angelo, I, Lin, L.Y, Dresen, C, Tocheva, E.I, Strynadka, N, Eltis, L.D. | | Deposit date: | 2010-02-28 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism
J.Biol.Chem., 285, 2010
|
|
1FC7
 
 | | PHOTOSYSTEM II D1 C-TERMINAL PROCESSING PROTEASE | | Descriptor: | PHOTOSYSTEM II D1 PROTEASE | | Authors: | Liao, D.I, Qian, J, Chisholm, D.A, Jordan, D.B, Diner, B.A. | | Deposit date: | 2000-07-18 | | Release date: | 2001-01-18 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the photosystem II D1 C-terminal processing protease.
Nat.Struct.Biol., 7, 2000
|
|
5C5U
 
 | | The crystal structure of viral collagen prolyl hydroxylase vCPH from Paramecium Bursaria Chlorella virus-1 - Truncated Construct | | Descriptor: | ACETATE ION, MANGANESE (II) ION, Prolyl 4-hydroxylase, ... | | Authors: | Longbotham, J.E, Levy, C.W, Johannisen, L.O, Tarhonskaya, H, Jiang, S, Loenarz, C, Flashman, E, Hay, S, Schofiled, C.J, Scrutton, N.S. | | Deposit date: | 2015-06-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Mechanism of a Viral Collagen Prolyl Hydroxylase.
Biochemistry, 54, 2015
|
|
5C66
 
 | |
8GO3
 
 | |
5C7O
 
 | | Structure of human testis-specific glyceraldehyde-3-phosphate dehydrogenase holo form with NAD+ | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Betts, L, Machius, M, Danshina, P, O'Brien, D. | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | Structural analyses to identify selective inhibitors of glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme.
Mol. Hum. Reprod., 22, 2016
|
|
1FG5
 
 | | CRYSTAL STRUCTURE OF BOVINE ALPHA-1,3-GALACTOSYLTRANSFERASE CATALYTIC DOMAIN. | | Descriptor: | N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE | | Authors: | Gastinel, L.N, Bignon, C, Shaper, J.H, Joziasse, D.H. | | Deposit date: | 2000-07-28 | | Release date: | 2001-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bovine alpha1,3-galactosyltransferase catalytic domain structure and its relationship with ABO histo-blood group and glycosphingolipid glycosyltransferases.
EMBO J., 20, 2001
|
|
6V18
 
 | | immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen beta, GLYCEROL, ... | | Authors: | Lim, J.J, Rossjohn, J. | | Deposit date: | 2019-11-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The shared susceptibility epitope of HLA-DR4 binds citrullinated self-antigens and the TCR.
Sci Immunol, 6, 2021
|
|
8GQ1
 
 | | HyHEL10 Fab complexed with hen egg lysozyme carrying arginine cluster in framework region of light chain. | | Descriptor: | Heavy Chain of HyHel10 Antibody Fragment (Fab), Light Chain of HyHel10 Antibody Fragment (Fab), Lysozyme C, ... | | Authors: | Matsuura, H, Hirata, K, Sakai, N, Nakakido, M, Tsumoto, K. | | Deposit date: | 2022-08-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Arginine cluster introduction on framework region in anti-lysozyme antibody improved association rate constant by changing conformational diversity of CDR loops.
Protein Sci., 32, 2023
|
|
6V38
 
 | | Cryo-EM structure of Ca2+-bound hsSlo1 channel | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Tao, X, MacKinnon, R. | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular structures of the human Slo1 K + channel in complex with beta 4.
Elife, 8, 2019
|
|
4QNQ
 
 | |
5CB3
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase | | Authors: | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
8GYK
 
 | | CryoEM structure of the RAD51_ADP filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA repair protein RAD51 homolog 1, MAGNESIUM ION | | Authors: | Miki, Y, Luo, S.C, Ho, M.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | A RAD51-ADP double filament structure unveils the mechanism of filament dynamics in homologous recombination.
Nat Commun, 14, 2023
|
|
1FJD
 
 | | HUMAN PARVULIN-LIKE PEPTIDYL PROLYL CIS/TRANS ISOMERASE, HPAR14 | | Descriptor: | PEPTIDYL PROLYL CIS/TRANS ISOMERASE (PPIASE) | | Authors: | Terada, T, Shirouzu, M, Fukumori, Y, Fujimori, F, Ito, Y, Kigawa, T, Yokoyama, S, Uchida, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human parvulin-like peptidyl prolyl cis/trans isomerase, hPar14.
J.Mol.Biol., 305, 2001
|
|
6V7D
 
 | | Human Arginase1 Complexed with Bicyclic Inhibitor Compound 10 | | Descriptor: | Arginase-1, MANGANESE (II) ION, {3-[(3aR,4R,5S,6aR)-4-azaniumyl-4-carboxyoctahydrocyclopenta[b]pyrrol-1-ium-5-yl]propyl}(trihydroxy)borate(1-) | | Authors: | Palte, R.L, Lesburg, C.A. | | Deposit date: | 2019-12-08 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery and Optimization of Rationally Designed Bicyclic Inhibitors of Human Arginase to Enhance Cancer Immunotherapy.
Acs Med.Chem.Lett., 11, 2020
|
|
6V51
 
 | | Spin-labeled T4 Lysozyme (9/131FnbY)-(4-Amino-TEMPO) | | Descriptor: | 4-amino-2,2,6,6-tetramethylpiperidin-1-ol, Endolysin | | Authors: | Liu, J, Morizumi, T, Ou, W.L, Wang, L, Ernst, O.P. | | Deposit date: | 2019-12-02 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Genetically Encoded Quinone Methides Enabling Rapid, Site-Specific, and Photocontrolled Protein Modification with Amine Reagents.
J.Am.Chem.Soc., 142, 2020
|
|
6V6L
 
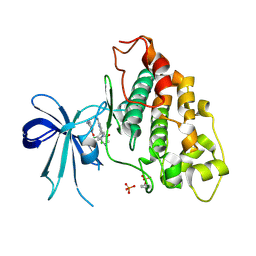 | | Co-structure of human glycogen synthase kinase beta with 1-(6-((2-((6-amino-5-nitropyridin-2-yl)amino)ethyl)amino)-2-(2,4-dichlorophenyl)pyridin-3-yl)-4-methylpiperazin-2-one | | Descriptor: | 1-(6-((2-((6-amino-5-nitropyridin-2-yl)amino)ethyl)amino)-2-(2,4-dichlorophenyl)pyridin-3-yl)-4-methylpiperazin-2-one, Glycogen synthase kinase-3 beta, PHOSPHATE ION | | Authors: | Bussiere, D.E, Fang, E, Shu, W. | | Deposit date: | 2019-12-05 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery and optimization of novel pyridines as highly potent and selective glycogen synthase kinase 3 inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6VAX
 
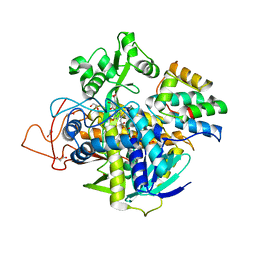 | | Crystal structure of human SDHA-SDHAF2 assembly intermediate | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Sharma, P, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2019-12-18 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The roles of SDHAF2 and dicarboxylate in covalent flavinylation of SDHA, the human complex II flavoprotein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1FGX
 
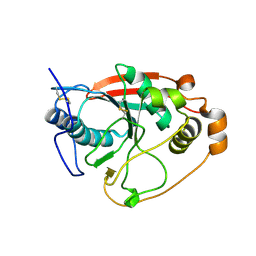 | | CRYSTAL STRUCTURE OF THE BOVINE BETA 1,4 GALACTOSYLTRANSFERASE (B4GALT1) CATALYTIC DOMAIN COMPLEXED WITH UMP | | Descriptor: | BETA 1,4 GALACTOSYLTRANSFERASE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Gastinel, L.N, Cambillau, C, Bourne, Y. | | Deposit date: | 2000-07-29 | | Release date: | 2000-08-16 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the bovine beta4galactosyltransferase catalytic domain and its complex with uridine diphosphogalactose.
EMBO J., 18, 1999
|
|
6VCM
 
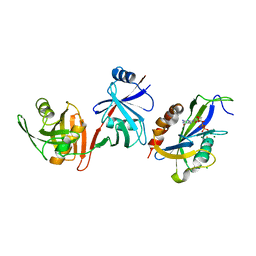 | | Crystal structure of E.coli RppH-DapF in complex with GTP, Mg2+ and F- | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, FLUORIDE ION, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
