8URT
 
 | | Cholinephosphotransferase in complex with selective inhibitor chelerythrine | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, Cholinephosphotransferase 1, ... | | Authors: | Roberts, J.R, Maeda, S, Ohi, M.D. | | Deposit date: | 2023-10-26 | | Release date: | 2024-10-30 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for catalysis and selectivity of phospholipid synthesis by eukaryotic choline-phosphotransferase.
Nat Commun, 16, 2025
|
|
7ZHY
 
 | |
7CRM
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-APO Structure | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside 2'-N-acetyltransferase | | Authors: | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2020-08-13 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
7ZHU
 
 | |
8QEZ
 
 | | Crystal structure of the AMPA receptor GluA2-L504Y-N775S ligand binding domain in complex with L-glutamate and positive allosteric modulator BPAM395 at 1.55A resolution | | Descriptor: | 6-chloranyl-4-cyclopropyl-2,3-dihydrothieno[3,2-e][1,2,4]thiadiazine 1,1-dioxide, ACETATE ION, CACODYLATE ION, ... | | Authors: | Dorosz, J, Laulumaa, S, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2023-09-01 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Exploring thienothiadiazine dioxides as isosteric analogues of benzo- and pyridothiadiazine dioxides in the search of new AMPA and kainate receptor positive allosteric modulators.
Eur.J.Med.Chem., 264, 2023
|
|
7ZHW
 
 | |
5I9U
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
6Q2T
 
 | | Human sterol 14a-demethylase (CYP51) in complex with the functionally irreversible inhibitor (R)-N-(1-(3-chloro-4'-fluoro-[1,1'-biphenyl]-4-yl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-(3-fluoro-5-(5-fluoropyrimidin-4-yl)phenyl)-1,3,4-oxadiazol-2-yl)benzamide | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, Lanosterol 14-alpha demethylase, N-[(1R)-1-(3-chloro-4'-fluoro[1,1'-biphenyl]-4-yl)-2-(1H-imidazol-1-yl)ethyl]-4-{5-[3-fluoro-5-(5-fluoropyrimidin-4-yl)phenyl]-1,3,4-oxadiazol-2-yl}benzamide, ... | | Authors: | Friggeri, L, Hargrove, T.Y, Wawrzak, Z, Lepesheva, G.I. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Validation of Human Sterol 14 alpha-Demethylase (CYP51) Druggability: Structure-Guided Design, Synthesis, and Evaluation of Stoichiometric, Functionally Irreversible Inhibitors.
J.Med.Chem., 62, 2019
|
|
7ZNV
 
 | | Artificial Unspecific Peroxygenase expressed in Pichia pastoris at 1.21 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Robinson, W.X.Q, Mielke, T, Grogan, G. | | Deposit date: | 2022-04-22 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Comparing the Catalytic and Structural Characteristics of a 'Short' Unspecific Peroxygenase (UPO) Expressed in Pichia pastoris and Escherichia coli.
Chembiochem, 24, 2023
|
|
9BK2
 
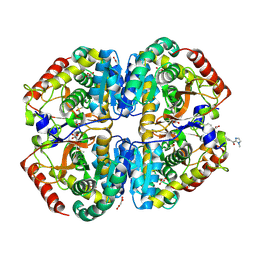 | | Crystal structure of Lactate dehydrogenase in complex with 4-((4-(1-methyl-1H-imidazole-2-carbonyl)phenyl)amino)-4-oxo-2-(4-(trifluoromethyl)phenyl)butanoic acid (S-enantiomer, monoclinic P form) | | Descriptor: | (2S)-4-[4-(1-methyl-1H-imidazole-2-carbonyl)anilino]-4-oxo-2-[4-(trifluoromethyl)phenyl]butanoic acid, DIMETHYL SULFOXIDE, L-lactate dehydrogenase A chain, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Sharma, H. | | Deposit date: | 2024-04-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and biological characterization of an orally bioavailable lactate dehydrogenase-A inhibitor against pancreatic cancer.
Eur.J.Med.Chem., 275, 2024
|
|
8XZZ
 
 | | Structure of a xylanase Xyl-1 M4 E175A in complex with xylobiose | | Descriptor: | Endo-1,4-beta-xylanase, ZINC ION, beta-D-xylopyranose, ... | | Authors: | Xiang, W.L, Huang, J.-W, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-01-21 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Unleashing the Power of Evolution in Xylanase Engineering: Investigating the Role of Distal Mutation Regulation.
J.Agric.Food Chem., 72, 2024
|
|
5MR3
 
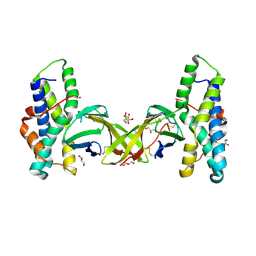 | | Crystal structure of red abalone egg VERL repeat 2 with linker in complex with sperm lysin at 1.8 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Egg-lysin, ... | | Authors: | Nishimura, K, Raj, I, Sadat Al-Hosseini, H, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
8VQQ
 
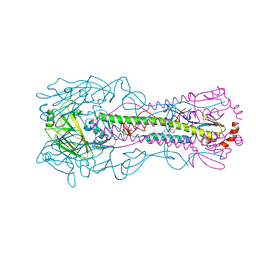 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule 6S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2024-01-19 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9C10
 
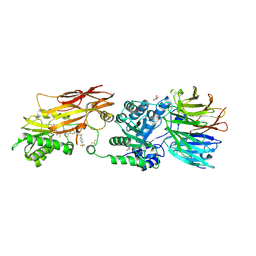 | | AMG 193, a clinical stage MTA-cooperative PRMT5 inhibitor, drives anti-tumor activity preclinically and in patients with MTAP-deleted cancers | | Descriptor: | (4-amino-1,3-dihydrofuro[3,4-c][1,7]naphthyridin-8-yl){(3S)-3-[4-(trifluoromethyl)phenyl]morpholin-4-yl}methanone, 5'-DEOXY-5'-METHYLTHIOADENOSINE, DIMETHYL SULFOXIDE, ... | | Authors: | Ghimire-Rijal, S, Mukund, S. | | Deposit date: | 2024-05-28 | | Release date: | 2024-10-02 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | AMG 193, a Clinical Stage MTA-Cooperative PRMT5 Inhibitor, Drives Antitumor Activity Preclinically and in Patients with MTAP-Deleted Cancers.
Cancer Discov, 15, 2025
|
|
8VQL
 
 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule 6S prime | | Descriptor: | (S~1~S,3S)-N-{3,5-dichloro-4-[(2S)-2-phenylmorpholine-4-carbonyl]phenyl}-3-(dimethylamino)pyrrolidine-1-sulfonimidoyl fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2024-01-18 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7LXL
 
 | | Crystal structure of human CYP3A4 bound to the testosterone dimer | | Descriptor: | 17alpha-hydroxy-7alpha-[(2Z)-4-(17beta-hydroxy-3-oxo-8alpha-androst-4-en-7beta-yl)but-2-en-1-yl]-8alpha,10alpha,13alpha,14beta-androst-4-en-3-one, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2021-03-04 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Innovative C 2 -symmetric testosterone and androstenedione dimers: Design, synthesis, biological evaluation on prostate cancer cell lines and binding study to recombinant CYP3A4.
Eur.J.Med.Chem., 220, 2021
|
|
6SFE
 
 | | CRYSTAL STRUCTURE OF DHQ1 FROM SALMONELLA TYPHI COVALENTLY MODIFIED BY COMPOUND 7 | | Descriptor: | (1~{S},3~{S},4~{S},5~{R})-3-(aminomethyl)-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Lence, E, Maneiro, M, Thompson, R, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Self-Immolation of a Bacterial Dehydratase Enzyme by its Epoxide Product.
Chemistry, 26, 2020
|
|
5IXY
 
 | | Lactate Dehydrogenase in complex with hydroxylactam inhibitor compound 31: (2~{S})-5-(2-chlorophenyl)sulfanyl-2-(4-morpholin-4-ylphenyl)-4-oxidanyl-2-thiophen-3-yl-1,3-dihydropyridin-6-one | | Descriptor: | (2~{S})-5-(2-chlorophenyl)sulfanyl-2-(4-morpholin-4-ylphenyl)-4-oxidanyl-2-thiophen-3-yl-1,3-dihydropyridin-6-one, L-lactate dehydrogenase A chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Chen, Z, Eigenbrot, C. | | Deposit date: | 2016-03-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cell Active Hydroxylactam Inhibitors of Human Lactate Dehydrogenase with Oral Bioavailability in Mice.
Acs Med.Chem.Lett., 7, 2016
|
|
6GMU
 
 | | Serum paraoxonase-1 by directed evolution with the L69G/H134R/F222S/T332S mutations | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ben-David, M, Sussman, J.L, Tawfik, D.S. | | Deposit date: | 2018-05-28 | | Release date: | 2019-04-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enzyme Evolution: An Epistatic Ratchet versus a Smooth Reversible Transition.
Mol.Biol.Evol., 37, 2020
|
|
5K30
 
 | | Crystal structure of methionine gamma-lyase from Citrobacter freundii modified by S-Ethyl-L-cysteine sulfoxide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Methionine gamma-lyase, ... | | Authors: | Revtovich, S.V, Nikulin, A.D, Morozova, E.A, Demidkina, T.V. | | Deposit date: | 2016-05-19 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Sulfoxides of sulfur-containing amino acids are suicide substrates of Citrobacter freundii methionine gamma-lyase. Structural bases of the enzyme inactivation.
Biochimie, 168, 2020
|
|
7X12
 
 | | Crystal structure of ME1 in complex with NADPH | | Descriptor: | MANGANESE (II) ION, NADP-dependent malic enzyme, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Amano, Y. | | Deposit date: | 2022-02-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery and Characterization of a Novel Allosteric Small-Molecule Inhibitor of NADP + -Dependent Malic Enzyme 1.
Biochemistry, 61, 2022
|
|
6DQ0
 
 | | sfGFP D133 mutated to 4-nitro-L-phenylalanine | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, superfolder green fluorescent protein | | Authors: | Phillips-Piro, C.M, Maurici, N, Lee, B. | | Deposit date: | 2018-06-10 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Crystal structures of green fluorescent protein with the unnatural amino acid 4-nitro-L-phenylalanine.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6Q6F
 
 | | Crystal structure of IDH1 R132H in complex with HMS101 | | Descriptor: | (2~{R})-2-[2-[(3~{R})-3-(4-fluorophenyl)pyrrolidin-1-yl]ethyl]-1,4-dimethyl-piperazine, Isocitrate dehydrogenase [NADP] cytoplasmic | | Authors: | Chaturvedi, A, Goparaju, R, Gupta, C, Kluenemann, T, Araujo Cruz, M.M, Kloos, A, Goerlich, K, Schottmann, R, Struys, E.A, Ganser, A, Preller, M, Heuser, M. | | Deposit date: | 2018-12-10 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | In vivo efficacy of mutant IDH1 inhibitor HMS-101 and structural resolution of distinct binding site.
Leukemia, 34, 2020
|
|
8XYG
 
 | | Crystal structure of SARS-CoV-2 BQ.1.1 RBD and human ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Lan, J, Wang, C.H. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Receptor binding mechanism and immune evasion capacity of SARS-CoV-2 BQ.1.1 lineage.
Virology, 600, 2024
|
|
9F64
 
 | | Crystal structure of thiol peroxidase mutant (C94A) in complex with Metro* (H. pylori | | Descriptor: | 1,2-ETHANEDIOL, 2-(5-azanyl-2-methyl-imidazol-1-yl)ethanol, SODIUM ION, ... | | Authors: | Fiedler, M.K, Gong, R, Fuchs, S, Rox, K, Friedrich, V, Pfeiffer, D, Reinhardt, T, Mibus, C, Huber, M, Hess, C, Mejias-Luque, R, Gerhard, M, Groll, M, Sieber, S.A. | | Deposit date: | 2024-04-30 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 5-Nitroimidazole ethers boost anti-Helicobacter pylori activity via a dual mode of action and effectively eradicate infections in vivo
to be published
|
|
