1ZCR
 
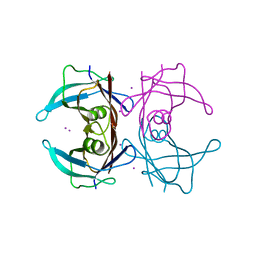 | | Crystal structure of human Transthyretin with bound iodide | | Descriptor: | IODIDE ION, Transthyretin | | Authors: | Hornberg, A, Hultdin, U.W, Olofsson, A, Sauer-Eriksson, A.E. | | Deposit date: | 2005-04-13 | | Release date: | 2005-07-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The effect of iodide and chloride on transthyretin structure and stability
Biochemistry, 44, 2005
|
|
6CK5
 
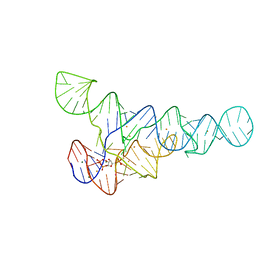 | | PRPP riboswitch from T. mathranii bound to PRPP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, BARIUM ION, MAGNESIUM ION, ... | | Authors: | Knappenberger, A.J, Reiss, C.W, Strobel, S.A. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structures of two aptamers with differing ligand specificity reveal ruggedness in the functional landscape of RNA.
Elife, 7, 2018
|
|
241D
 
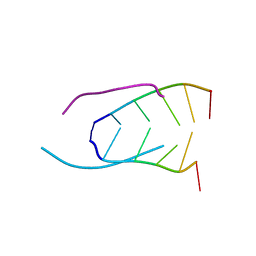 | | EXTENSION OF THE FOUR-STRANDED INTERCALATED CYTOSINE MOTIF BY ADENINE.ADENINE BASE PAIRING IN THE CRYSTAL STRUCTURE OF D(CCCAAT) | | Descriptor: | DNA (5'-D(*CP*CP*CP*AP*AP*T)-3') | | Authors: | Berger, I, Kang, C, Fredian, A, Ratliff, R, Moyzis, R, Rich, A. | | Deposit date: | 1995-12-07 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Extension of the four-stranded intercalated cytosine motif by adenine.adenine base pairing in the crystal structure of d(CCCAAT).
Nat.Struct.Biol., 2, 1995
|
|
1ZEM
 
 | |
1YZD
 
 | |
1GPQ
 
 | | Structure of ivy complexed with its target, HEWL | | Descriptor: | INHIBITOR OF VERTEBRATE LYSOZYME, LYSOZYME C | | Authors: | Abergel, C, Monchois, V, Claverie, J.-M. | | Deposit date: | 2001-11-08 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Evolution of the Ivy Protein Family, Unexpected Lysozyme Inhibitors in Gram-Negative Bacteria.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1T4F
 
 | | Structure of human MDM2 in complex with an optimized p53 peptide | | Descriptor: | SULFATE ION, Ubiquitin-protein ligase E3 Mdm2, optimized p53 peptide | | Authors: | Grasberger, B.L, Schubert, C, Koblish, H.K, Carver, T.E, Franks, C.F, Zhao, S.Y, Lu, T, LaFrance, L.V, Parks, D.J. | | Deposit date: | 2004-04-29 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells
J.Med.Chem., 48, 2005
|
|
6RGI
 
 | | Partially unfolded cytochrome c in complex with sulfonatocalix[6]arene | | Descriptor: | Cytochrome c iso-1, HEME C, IMIDAZOLE, ... | | Authors: | Engilberge, S, Rennie, M.L, Crowley, P.B. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Calixarene capture of partially unfolded cytochrome c.
Febs Lett., 593, 2019
|
|
1RC8
 
 | | T4 Polynucleotide Kinase bound to 5'-GTCAC-3' ssDNA | | Descriptor: | 5'-D(*GP*TP*CP*AP*C)-3', ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Eastberg, J.H, Pelletier, J, Stoddard, B.L. | | Deposit date: | 2003-11-03 | | Release date: | 2004-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Recognition of DNA substrates by T4 bacteriophage polynucleotide kinase.
Nucleic Acids Res., 32, 2004
|
|
1U5S
 
 | | NMR structure of the complex between Nck-2 SH3 domain and PINCH-1 LIM4 domain | | Descriptor: | Cytoplasmic protein NCK2, PINCH protein, ZINC ION | | Authors: | Vaynberg, J, Fukuda, T, Vinogradova, O, Velyvis, A, Ng, L, Wu, C, Qin, J. | | Deposit date: | 2004-07-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an ultraweak protein-protein complex and its crucial role in regulation of cell morphology and motility.
Mol.Cell, 17, 2005
|
|
6RSL
 
 | |
6RSK
 
 | |
6RSI
 
 | |
1U6O
 
 | | Mispairing of a Site-Specific Major Groove (2S,3S)-N6-(2,3,4-Trihydroxybutyl)-2-deoxyadenosyl DNA Adduct of Butadiene Diol Epoxide with Deoxyguanosine: Formation of a dA(anti)dG(anti) Pairing Interaction | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(2BU)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*GP*GP*TP*CP*CP*G)-3' | | Authors: | Scholdberg, T.A, Nechev, L.N, Merritt, W.K, Harris, T.M, Harris, C.M, Lloyd, R.S, Stone, M.P. | | Deposit date: | 2004-07-30 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mispairing of a Site Specific Major Groove (2S,3S)-N6-(2,3,4-Trihydroxybutyl)-2'-deoxyadenosyl DNA Adduct of Butadiene Diol Epoxide with Deoxyguanosine: Formation of a dA(anti)dG(anti) Pairing Interaction.
CHEM.RES.TOXICOL., 18, 2005
|
|
1U6G
 
 | | Crystal Structure of The Cand1-Cul1-Roc1 Complex | | Descriptor: | Cullin homolog 1, RING-box protein 1, TIP120 protein, ... | | Authors: | Goldenberg, S.J, Shumway, S.D, Cascio, T.C, Garbutt, K.C, Liu, J, Xiong, Y, Zheng, N. | | Deposit date: | 2004-07-29 | | Release date: | 2004-12-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases
Cell(Cambridge,Mass.), 119, 2004
|
|
6SBU
 
 | | X-ray Structure of Human LDHA with an Allosteric Inhibitor (Compound 3) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-[[4-[(5-chloranylthiophen-2-yl)carbonylamino]-1,3-bis(oxidanylidene)isoindol-2-yl]methyl]benzoic acid, L-lactate dehydrogenase A chain | | Authors: | Friberg, A, Puetter, V, Nguyen, D, Rehwinkel, H. | | Deposit date: | 2019-07-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural Evidence for Isoform-Selective Allosteric Inhibition of Lactate Dehydrogenase A.
Acs Omega, 5, 2020
|
|
6SBV
 
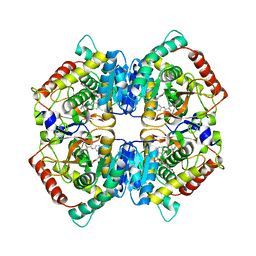 | | X-ray Structure of Human LDH-A with an Allosteric Inhibitor (Compound 7) | | Descriptor: | L-lactate dehydrogenase A chain, ~{N}-[3-[(7-nitrodibenzofuran-2-yl)sulfonylamino]phenyl]-1-oxidanyl-cyclopropane-1-carboxamide | | Authors: | Friberg, A, Puetter, V, Nguyen, D, Rehwinkel, H. | | Deposit date: | 2019-07-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Evidence for Isoform-Selective Allosteric Inhibition of Lactate Dehydrogenase A.
Acs Omega, 5, 2020
|
|
1U6N
 
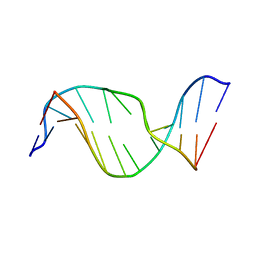 | | Solution Structure of an Oligodeoxynucleotide Containing a Butadiene Derived N1 b-Hydroxyalkyl Adduct on Deoxyinosine in the Human N-ras Codon 61 Sequence | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(2BD)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Scholdberg, T.A, Merritt, W.K, Dean, S.M, Kowalcyzk, A, Harris, T.M, Harris, C.M, Lloyd, R.S, Stone, M.P. | | Deposit date: | 2004-07-30 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an Oligodeoxynucleotide Containing a Butadiene Oxide-Derived N1 Beta-Hydroxyalkyl Deoxyinosine Adduct in the Human N-ras Codon 61 Sequence.
Biochemistry, 44, 2005
|
|
1RPZ
 
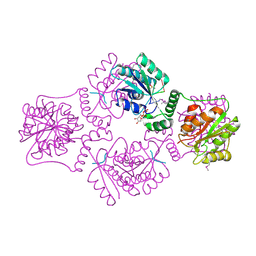 | | T4 POLYNUCLEOTIDE KINASE BOUND TO 5'-TGCAC-3' SSDNA | | Descriptor: | 5'-D(*TP*GP*CP*AP*C)-3', ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Eastberg, J.H, Pelletier, J, Stoddard, B.L. | | Deposit date: | 2003-12-03 | | Release date: | 2004-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition of DNA substrates by T4 bacteriophage polynucleotide kinase.
Nucleic Acids Res., 32, 2004
|
|
1CTZ
 
 | |
3ZOU
 
 | | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PA01, with bound fragment SPB02696, and substrate geranyl pyrophosphate. | | Descriptor: | 3-(2-oxo-1,3-benzoxazol-3(2H)-yl)propanoic acid, DIMETHYL SULFOXIDE, FARNESYL PYROPHOSPHATE SYNTHASE, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2013-02-25 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Characterization of Substrate and Inhibitor Binding to Farnesyl Pyrophosphate Synthase from Pseudomonas Aeruginosa
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3ZMC
 
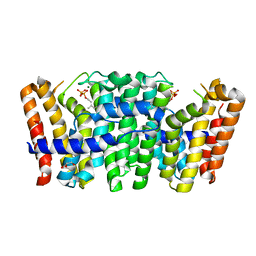 | | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PA01, with bound substrate molecule Geranyl pyrophosphate. | | Descriptor: | DIMETHYL SULFOXIDE, GERANYL DIPHOSPHATE, GERANYLTRANSTRANSFERASE, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2013-02-07 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Characterization of Substrate and Inhibitor Binding to Farnesyl Pyrophosphate Synthase from Pseudomonas Aeruginosa
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1CTY
 
 | |
1CRZ
 
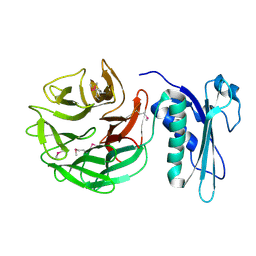 | | CRYSTAL STRUCTURE OF THE E. COLI TOLB PROTEIN | | Descriptor: | TOLB PROTEIN | | Authors: | Abergel, C, Bouveret, E, Claverie, J.-M, Brown, K, Rigal, A, Lazdunski, C, Benedetti, H. | | Deposit date: | 1999-08-16 | | Release date: | 2000-08-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Escherichia coli TolB protein determined by MAD methods at 1.95 A resolution.
Structure Fold.Des., 7, 1999
|
|
3ZCD
 
 | |
