3MUN
 
 | | APPEP_PEPCLOSE closed state | | Descriptor: | GLYCEROL, Prolyl endopeptidase, SULFATE ION, ... | | Authors: | Chiu, T.K. | | Deposit date: | 2010-05-03 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Route of Substrate Entry in Prolyl Endopeptidase
TO BE PUBLISHED
|
|
3MUO
 
 | | APPEP_PEPCLOSE+PP closed state | | Descriptor: | GLYCEROL, N-BENZYLOXYCARBONYL-L-PROLYL-L-PROLINAL, Prolyl endopeptidase, ... | | Authors: | Chiu, T.K. | | Deposit date: | 2010-05-03 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Route of Substrate Entry in Prolyl Endopeptidase
TO BE PUBLISHED
|
|
3OBV
 
 | | Autoinhibited Formin mDia1 Structure | | Descriptor: | Protein diaphanous homolog 1, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Tomchick, D.R, Rosen, M.K, Otomo, T. | | Deposit date: | 2010-08-09 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the Formin mDia1 in autoinhibited conformation.
Plos One, 5, 2010
|
|
3OVN
 
 | | Fragment-based approach to the design of ligands targeting a novel site on HIV-1 integrase | | Descriptor: | 1-methyl-3-(thiophen-2-yl)-1H-pyrazol-5-amine, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Headey, S.J, Deadman, J.J, Rhodes, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2010-09-16 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1
Chemmedchem, 6, 2011
|
|
3Q73
 
 | | Cryptococcus neoformans protein farnesyltransferase, apo enzyme | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, Farnesyltransferase, alpha subunit, ... | | Authors: | Hast, M.A, Beese, L.S. | | Deposit date: | 2011-01-04 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Cryptococcus neoformans Protein Farnesyltransferase Reveal Strategies for Developing Inhibitors That Target Fungal Pathogens.
J.Biol.Chem., 286, 2011
|
|
3Q7A
 
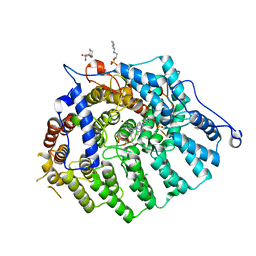 | | Cryptococcus neoformans protein farnesyltransferase in complex with FPP and L-778,123 | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 4-[(5-{[4-(3-CHLOROPHENYL)-3-OXOPIPERAZIN-1-YL]METHYL}-1H-IMIDAZOL-1-YL)METHYL]BENZONITRILE, FARNESYL DIPHOSPHATE, ... | | Authors: | Hast, M.A, Beese, L.S. | | Deposit date: | 2011-01-04 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Cryptococcus neoformans Protein Farnesyltransferase Reveal Strategies for Developing Inhibitors That Target Fungal Pathogens.
J.Biol.Chem., 286, 2011
|
|
3Q7F
 
 | |
3QGV
 
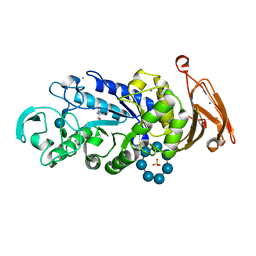 | | Crystal structure of a thermostable amylase variant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha amylase, CALCIUM ION, ... | | Authors: | Hein, K.L, Ganshaw, G, Bott, R, Nissen, P. | | Deposit date: | 2011-01-25 | | Release date: | 2012-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a highly mutated, thermostable \xCE\xB1-amylase variant
To be Published
|
|
3QPH
 
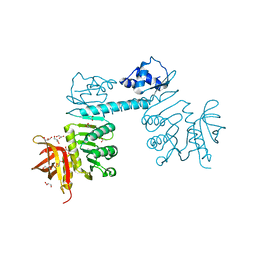 | | The three-dimensional structure of TrmB, a global transcriptional regulator of the hyperthermophilic archaeon Pyrococcus furiosus in complex with sucrose | | Descriptor: | ACETATE ION, GLYCEROL, TrmB, ... | | Authors: | Krug, M, Lee, S.-J, Boos, W, Welte, W, Diederichs, K. | | Deposit date: | 2011-02-13 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.992 Å) | | Cite: | The three-dimensional structure of TrmB, a transcriptional regulator of dual function in the hyperthermophilic archaeon Pyrococcus furiosus in complex with sucrose.
Protein Sci., 22, 2013
|
|
3QYQ
 
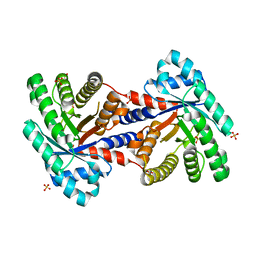 | | 1.8 Angstrom resolution crystal structure of a putative deoxyribose-phosphate aldolase from Toxoplasma gondii ME49 | | Descriptor: | Deoxyribose-phosphate aldolase, putative, SULFATE ION, ... | | Authors: | Halavaty, A.S, Ruan, J, Minasov, G, Shuvalova, L, Ueno, A, Igarashi, M, Ngo, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional divergence of the aldolase fold in Toxoplasma gondii.
J.Mol.Biol., 427, 2015
|
|
3R1B
 
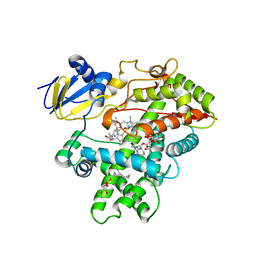 | | Open crystal structure of cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene | | Descriptor: | (4-tert-butylphenyl)acetaldehyde, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | Authors: | Gay, S.C, Zhang, H, Stout, C.D, Hollenberg, P.F, Halpert, J.R. | | Deposit date: | 2011-03-09 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Analysis of Mammalian Cytochrome P450 2B4 Covalently Bound to the Mechanism-Based Inactivator tert-Butylphenylacetylene: Insight into Partial Enzymatic Activity.
Biochemistry, 50, 2011
|
|
3S7Z
 
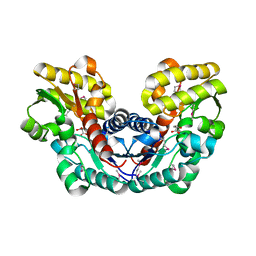 | | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium Complexed with Succinate | | Descriptor: | MAGNESIUM ION, Putative aspartate racemase, SUCCINIC ACID, ... | | Authors: | Maltseva, N, Zhang, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-27 | | Release date: | 2011-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium Complexed with Succinate.
To be Published
|
|
3SO3
 
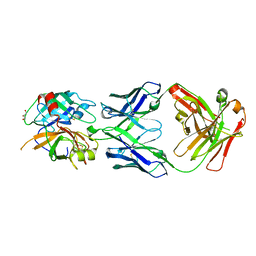 | | Structures of Fab-Protease Complexes Reveal a Highly Specific Non-Canonical Mechanism of Inhibition. | | Descriptor: | A11 FAB heavy chain, A11 FAB light chain, GLYCEROL, ... | | Authors: | Schneider, E.L, Farady, C.J, Egea, P.F, Goetz, D.H, Baharuddin, A, Craik, C.S. | | Deposit date: | 2011-06-29 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A reverse binding motif that contributes to specific protease inhibition by antibodies.
J.Mol.Biol., 415, 2012
|
|
3SP3
 
 | | Lysozyme in 20% sucrose | | Descriptor: | CHLORIDE ION, Lysozyme, SODIUM ION, ... | | Authors: | Sharma, P, Solanki, A.K, Ashish | | Deposit date: | 2011-07-01 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Lysozyme in 20% sucrose
to be published
|
|
3T3Z
 
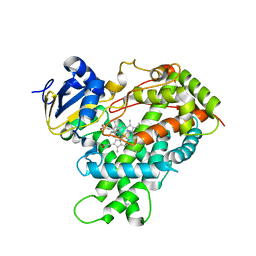 | | Human Cytochrome P450 2E1 in complex with pilocarpine | | Descriptor: | (3S,4R)-3-ethyl-4-[(1-methyl-1H-imidazol-5-yl)methyl]dihydrofuran-2(3H)-one, Cytochrome P450 2E1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Meneely, K.M, DeVore, N.M, Scott, E.E. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine.
Febs J., 279, 2012
|
|
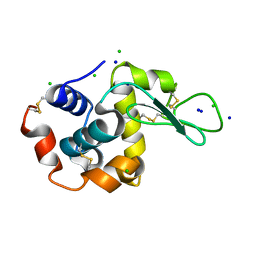
3T6U
 
 | |
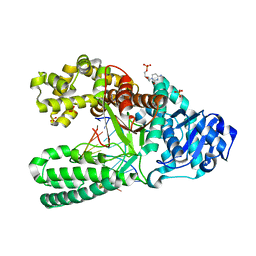
3TAN
 
 | |
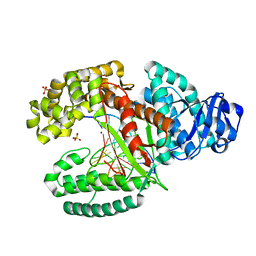
3TAP
 
 | |
3TAQ
 
 | |
3TAR
 
 | |
3VF1
 
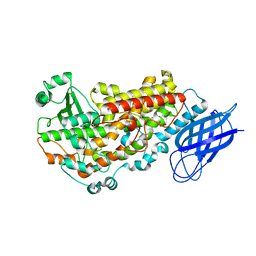 | | Structure of a calcium-dependent 11R-lipoxygenase suggests a mechanism for Ca-regulation | | Descriptor: | 11R-lipoxygenase, FE (II) ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Eek, P, Jarving, R, Jarving, I, Gilbert, N.C, Newcomer, M.E, Samel, N. | | Deposit date: | 2012-01-09 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | Structure of a Calcium-dependent 11R-Lipoxygenase Suggests a Mechanism for Ca2+ Regulation.
J.Biol.Chem., 287, 2012
|
|
3VQ7
 
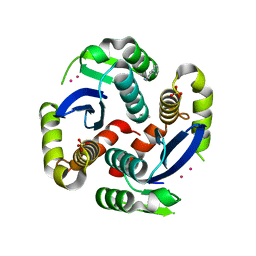 | | HIV-1 IN core domain in complex with 4-(1H-pyrrol-1-yl)aniline | | Descriptor: | 4-(1H-pyrrol-1-yl)aniline, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
3VQA
 
 | | HIV-1 IN core domain in complex with 1-benzothiophen-6-amine 1,1-dioxide | | Descriptor: | 1-benzothiophen-6-amine 1,1-dioxide, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
3VQQ
 
 | | HIV-1 integrase core domain in complex with 2,1,3-benzothiadiazol-4-amine | | Descriptor: | 2,1,3-benzothiadiazol-4-amine, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-29 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
4B9L
 
 | | Structure of the high fidelity DNA polymerase I with the oxidative formamidopyrimidine-dA DNA lesion in the pre-insertion site. | | Descriptor: | 5'-D(*CP*AP*GP*FAX*AP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
