3AEK
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N, ... | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3AEU
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3AER
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
1P90
 
 | | The Three-dimensional Structure of the Core Domain of NafY from Azotobacter vinelandii determined at 1.8 resolution | | Descriptor: | 1,2-ETHANEDIOL, hypothetical protein | | Authors: | Dyer, D.H, Rubio, L.M, Thoden, J.B, Holden, H.M, Ludden, P.W, Rayment, I. | | Deposit date: | 2003-05-08 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Three-dimensional Structure of the Core Domain of NafY from Azotobacter vinelandii determined at 1.8 A resolution
J.Biol.Chem., 278, 2003
|
|
5K9B
 
 | | Azotobacter vinelandii Flavodoxin II | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin-2, MAGNESIUM ION | | Authors: | Rees, D.C, Segal, H.M, Spatzal, T. | | Deposit date: | 2016-05-31 | | Release date: | 2017-08-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.174 Å) | | Cite: | Electrochemical and structural characterization of Azotobacter vinelandii flavodoxin II.
Protein Sci., 26, 2017
|
|
5V5W
 
 | |
5V5V
 
 | | Complex of NLGN2 with MDGA1 Ig1-Ig2 | | Descriptor: | MAM domain-containing glycosylphosphatidylinositol anchor protein 1, Neuroligin-2 | | Authors: | Gangwar, S.P, Machius, M, Rudenko, G. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.11 Å) | | Cite: | Molecular Mechanism of MDGA1: Regulation of Neuroligin 2:Neurexin Trans-synaptic Bridges.
Neuron, 94, 2017
|
|
5OJ6
 
 | | Crystal structure of the chicken MDGA1 ectodomain in complex with the human neuroligin 1 (NL1(-A-B)) cholinesterase domain. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAM domain-containing glycosylphosphatidylinositol anchor protein 1, Neuroligin-1, ... | | Authors: | Elegheert, J, Clayton, A.J, Aricescu, A.R. | | Deposit date: | 2017-07-20 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Mechanism for Modulation of Synaptic Neuroligin-Neurexin Signaling by MDGA Proteins.
Neuron, 95, 2017
|
|
5OJ2
 
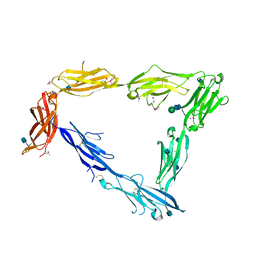 | | Crystal structure of the chicken MDGA1 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAM domain-containing glycosylphosphatidylinositol anchor protein 1, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Elegheert, J, Clayton, A.J, Aricescu, A.R. | | Deposit date: | 2017-07-20 | | Release date: | 2017-08-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Mechanism for Modulation of Synaptic Neuroligin-Neurexin Signaling by MDGA Proteins.
Neuron, 95, 2017
|
|
5XEQ
 
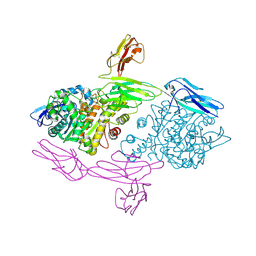 | | Crystal Structure of human MDGA1 and human neuroligin-2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAM domain-containing glycosylphosphatidylinositol anchor protein 1, ... | | Authors: | Kim, H.M, Kim, J.A, Kim, D. | | Deposit date: | 2017-04-05 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.136 Å) | | Cite: | Structural Insights into Modulation of Neurexin-Neuroligin Trans-synaptic Adhesion by MDGA1/Neuroligin-2 Complex
Neuron, 94, 2017
|
|
3ZYY
 
 | | Reductive activator for corrinoid,iron-sulfur protein | | Descriptor: | (R,R)-2,3-BUTANEDIOL, FE2/S2 (INORGANIC) CLUSTER, IRON-SULFUR CLUSTER BINDING PROTEIN, ... | | Authors: | Hennig, S.E, Jeoung, J.-H, Goetzl, S, Dobbek, H. | | Deposit date: | 2011-08-30 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Redox-Dependent Complex Formation by an ATP-Dependent Activator of the Corrinoid/Iron-Sulfur Protein.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2WC1
 
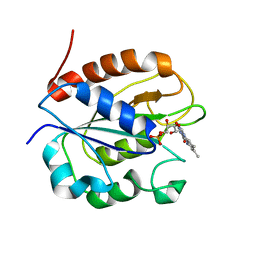 | | Three-dimensional Structure of the Nitrogen Fixation Flavodoxin (NifF) from Rhodobacter capsulatus at 2.2 A | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Perez-Dorado, I, Bittel, C, Hermoso, J.A, Cortez, N, Carrillo, N. | | Deposit date: | 2009-03-06 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural and Phylogenetic Analysis of Rhodobacter Capsulatus Niff: Uncovering General Features of Nitrogen-Fixation (Nif)-Flavodoxins.
Int.J.Mol.Sci., 14, 2013
|
|
6Y01
 
 | |
8AHX
 
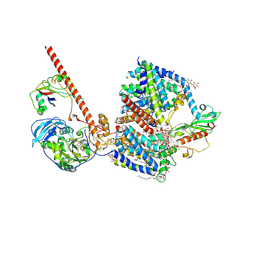 | |
4F6T
 
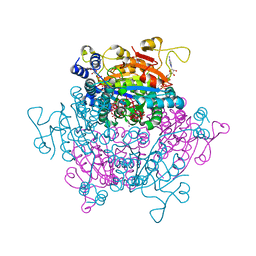 | | The crystal structure of the molybdenum storage protein (MoSto) from Azotobacter vinelandii loaded with various polyoxometalates | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MO(6)-O(26) Cluster, ... | | Authors: | Kowalewski, B, Poppe, J, Schneider, K, Demmer, U, Warkentin, E, Ermler, U. | | Deposit date: | 2012-05-15 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nature's Polyoxometalate Chemistry: X-ray Structure of the Mo Storage Protein Loaded with Discrete Polynuclear Mo-O Clusters.
J.Am.Chem.Soc., 134, 2012
|
|
3SBP
 
 | | Pseudomonas stutzeri nitrous oxide reductase, P1 crystal form | | Descriptor: | CALCIUM ION, CHLORIDE ION, DINUCLEAR COPPER ION, ... | | Authors: | Pomowski, A, Zumft, W.G, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2011-06-06 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | N2O binding at a [4Cu:2S] copper-sulphur cluster in nitrous oxide reductase.
Nature, 477, 2011
|
|
3SBR
 
 | | Pseudomonas stutzeri nitrous oxide reductase, P1 crystal form with substrate | | Descriptor: | CALCIUM ION, CHLORIDE ION, DINUCLEAR COPPER ION, ... | | Authors: | Pomowski, A, Zumft, W.G, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2011-06-06 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | N2O binding at a [4Cu:2S] copper-sulphur cluster in nitrous oxide reductase.
Nature, 477, 2011
|
|
3SBQ
 
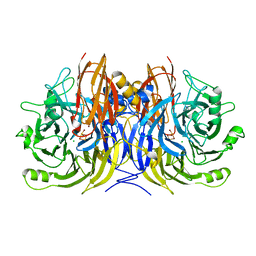 | | Pseudomonas stutzeri nitrous oxide reductase, P65 crystal form | | Descriptor: | CALCIUM ION, CHLORIDE ION, DINUCLEAR COPPER ION, ... | | Authors: | Pomowski, A, Zumft, W.G, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2011-06-06 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | N2O binding at a [4Cu:2S] copper-sulphur cluster in nitrous oxide reductase.
Nature, 477, 2011
|
|
8RB8
 
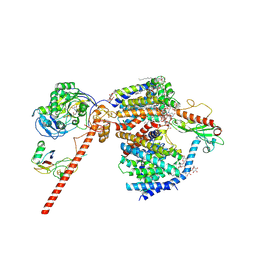 | |
8RB9
 
 | |
8RBM
 
 | |
8RBQ
 
 | |
1UUX
 
 | | Structure of a molybdopterin-bound cnx1g domain links molybdenum and copper metabolism | | Descriptor: | COPPER (I) ION, FORMIC ACID, IMIDAZOLE, ... | | Authors: | Kuper, J, Llamas, A, Hecht, H.J, Mendel, R.R, Schwarz, G. | | Deposit date: | 2004-01-12 | | Release date: | 2004-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a Molybdopterin-Bound Cnx1G Domain Links Molybdenum and Copper Metabolism
Nature, 430, 2004
|
|
1UUY
 
 | | Structure of a molybdopterin-bound cnx1g domain links molybdenum and copper metabolism | | Descriptor: | ADENOSINE MONOPHOSPHATE, COPPER (I) ION, FORMIC ACID, ... | | Authors: | Kuper, J, Llamas, A, Hecht, H.J, Mendel, R.R, Schwarz, G. | | Deposit date: | 2004-01-12 | | Release date: | 2004-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a Molybdopterin-Bound Cnx1G Domain Links Molybdenum and Copper Metabolism
Nature, 430, 2004
|
|
6UYK
 
 | | Dark-operative protochlorophyllide oxidoreductase in the nucleotide-free form. | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase iron-sulfur ATP-binding protein | | Authors: | Bacik, J.P, Imran, S.M.S, Watkins, M.B, Corless, E, Antony, E, Ando, N. | | Deposit date: | 2019-11-13 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The flexible N-terminus of BchL autoinhibits activity through interaction with its [4Fe-4S] cluster and released upon ATP binding.
J.Biol.Chem., 296, 2020
|
|
