8R2B
 
 | |
6Z71
 
 | | Structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2020-05-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of the Aquifex aeolicus MATE family multidrug resistance transporter and sequence comparisons suggest the existence of a new subfamily.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6Z70
 
 | | Structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2020-05-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the Aquifex aeolicus MATE family multidrug resistance transporter and sequence comparisons suggest the existence of a new subfamily.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8THI
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (parallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, SODIUM ION, Sialic acid TRAP transporter permease protein SiaT | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
2QA2
 
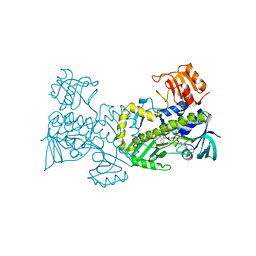 | | Crystal structure of CabE, an aromatic hydroxylase from angucycline biosynthesis, determined to 2.7 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyketide oxygenase CabE | | Authors: | Koskiniemi, H, Dobritzsch, D, Metsa-Ketela, M, Kallio, P, Niemi, J, Schneider, G. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of two aromatic hydroxylases involved in the early tailoring steps of angucycline biosynthesis
J.Mol.Biol., 372, 2007
|
|
4DNO
 
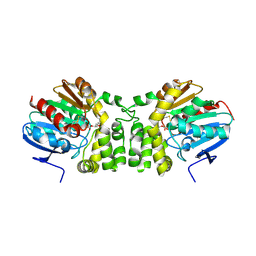 | | Crystal structure of the CFTR inhibitory factor Cif with the E153Q mutation adducted with the 1,2-epoxyhexane hydrolysis intermediate | | Descriptor: | (2R)-hexane-1,2-diol, Putative hydrolase | | Authors: | Bahl, C.D, Patankar, Y.R, Madden, D.R. | | Deposit date: | 2012-02-08 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Visualizing the Mechanism of Epoxide Hydrolysis by the Bacterial Virulence Enzyme Cif.
Biochemistry, 55, 2016
|
|
4DNF
 
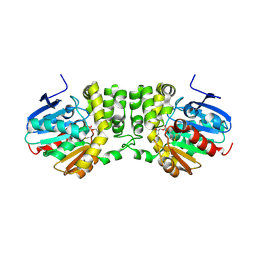 | |
4DM7
 
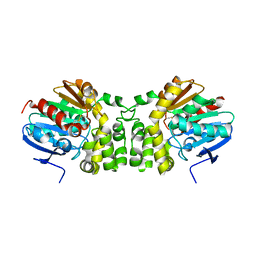 | |
4DLN
 
 | |
2QLH
 
 | | mPlum I65L mutant | | Descriptor: | Fluorescent protein plum | | Authors: | Shu, X, Remington, S.J. | | Deposit date: | 2007-07-12 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies of Far-Red Emission in mPlum, a Monomeric Red Fluorescent Protein
To be Published
|
|
8AW5
 
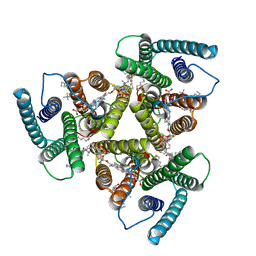 | | Cryo-EM structure of heme A synthase trimer from Aquifex aeolicus | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Heme O oxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hui, Z, Guoliang, Z. | | Deposit date: | 2022-08-29 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of heme A synthase trimer from Aquifex aeolicus
To Be Published
|
|
2QAR
 
 | |
2QLG
 
 | | mPlum | | Descriptor: | Fluorescent protein plum | | Authors: | Shu, X, Remington, S.J. | | Deposit date: | 2007-07-12 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies of Far-Red Emission in mPlum, a Monomeric Red Fluorescent Protein
To be Published
|
|
4Y1S
 
 | | Structural basis for Ca2+-mediated interaction of the perforin C2 domain with lipid membranes | | Descriptor: | CALCIUM ION, Perforin-1 | | Authors: | Conroy, P.J, Yagi, H, Whisstock, J.C, Norton, R.S. | | Deposit date: | 2015-02-09 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Structural Basis for Ca2+-mediated Interaction of the Perforin C2 Domain with Lipid Membranes.
J.Biol.Chem., 290, 2015
|
|
4EX6
 
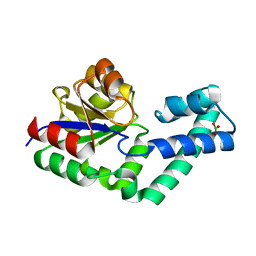 | | Crystal structure of the alnumycin P phosphatase AlnB | | Descriptor: | AlnB, BORIC ACID, MAGNESIUM ION | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2QLI
 
 | | mPlum E16Q mutant | | Descriptor: | Fluorescent protein plum | | Authors: | Shu, X, Remington, S.J. | | Deposit date: | 2007-07-12 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural Studies of Far-Red Emission in mPlum, a Monomeric Red Fluorescent Protein
To be Published
|
|
4Y1T
 
 | | Structural basis for Ca2+-mediated interaction of the perforin C2 domain with lipid membranes | | Descriptor: | CALCIUM ION, Perforin-1 | | Authors: | Conroy, P.J, Yagi, H, Whisstock, J.C, Norton, R.S. | | Deposit date: | 2015-02-09 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.666 Å) | | Cite: | Structural Basis for Ca2+-mediated Interaction of the Perforin C2 Domain with Lipid Membranes.
J.Biol.Chem., 290, 2015
|
|
8BAP
 
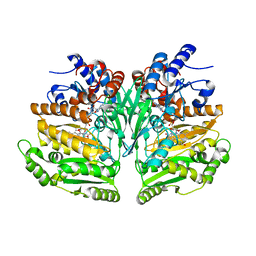 | | Eugenol Oxidase (EUGO) from Rhodococcus jostii RHA1, eightfold mutant active on propanol syringol | | Descriptor: | 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2,6-dimethoxyphenol, CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alvigini, L, Mattevi, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | One-Pot Biocatalytic Synthesis of rac -Syringaresinol from a Lignin-Derived Phenol.
Acs Catalysis, 13, 2023
|
|
4YX9
 
 | |
8C6Z
 
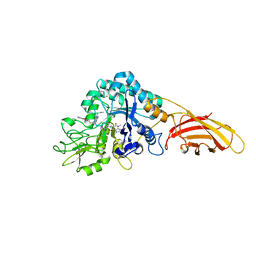 | | necrotic enteritis associated Clostridium p. chitinase F8UNI4 in complex with inhibitor bisdionin C | | Descriptor: | 1,1'-PROPANE-1,3-DIYLBIS(3,7-DIMETHYL-3,7-DIHYDRO-1H-PURINE-2,6-DIONE), CADMIUM ION, COBALT (II) ION, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Contribution and structural characterization of Chitinases to Clostridium perfringens Pathogenesis in Broilers
To Be Published
|
|
4ZEJ
 
 | |
5AC3
 
 | | Crystal structure of PAM12A | | Descriptor: | ACETIC ACID, CADMIUM ION, PEPTIDE AMIDASE | | Authors: | Wu, B, Wijma, H.J, Song, L, Rozeboom, H.J, Poloni, C, Tian, Y, Arif, M.I, Nuijens, T, Quadflieg, P.J.L.M, Szymanski, W, Feringa, B.L, Janssen, D.B. | | Deposit date: | 2015-08-11 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Versatile Peptide C-Terminal Functionalization Via a Computationally Peptide Amidase
Acs Catalysis, 2016
|
|
4O29
 
 | |
4PFE
 
 | | Crystal structure of vsfGFP-0 | | Descriptor: | Green fluorescent protein | | Authors: | Jauch, R, Chen, S.L. | | Deposit date: | 2014-04-29 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Rational Structure-Based Design of Bright GFP-Based Complexes with Tunable Dimerization.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4OSP
 
 | | The crystal structure of urdamycin C-6 ketoreductase domain UrdMred with bound NADP and rabelomycin | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxygenase-reductase, rabelomycin | | Authors: | Niiranen, L, Paananen, P, Patrikainen, P, Metsa-Ketela, M. | | Deposit date: | 2014-02-13 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based engineering of angucyclinone 6-ketoreductases.
Chem.Biol., 21, 2014
|
|
