4J9C
 
 | |
1AEY
 
 | |
1B07
 
 | | CRK SH3 DOMAIN COMPLEXED WITH PEPTOID INHIBITOR | | Descriptor: | PHENYLETHANE, PROTEIN (PROTO-ONCOGENE CRK (CRK)), PROTEIN (SH3 PEPTOID INHIBITOR) | | Authors: | Nguyen, J.T, Turck, C.W, Cohen, F.E, Zuckermann, R.N, Lim, W.A. | | Deposit date: | 1998-11-17 | | Release date: | 1999-01-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Exploiting the basis of proline recognition by SH3 and WW domains: design of N-substituted inhibitors.
Science, 282, 1998
|
|
1AWO
 
 | |
4JJD
 
 | |
4JJC
 
 | |
2RNA
 
 | | Itk SH3 average minimized | | Descriptor: | Tyrosine-protein kinase ITK/TSK | | Authors: | Severin, A.J. | | Deposit date: | 2007-12-08 | | Release date: | 2007-12-25 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure note: murine Itk SH3 domain
To be Published
|
|
2RQV
 
 | |
2RMO
 
 | | Solution structure of alpha-spectrin_SH3-bergerac from Chicken | | Descriptor: | Spectrin alpha chain, brain | | Authors: | Kutyshenko, V.P, Prokhorov, D.A, Timchenko, M.A, Kudrevatykh, Y.A, Fedyukina, D.V, Gushchina, L.V, Khristoforov, V.S, Filimonov, V.V. | | Deposit date: | 2007-11-07 | | Release date: | 2008-09-30 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Study of the structure and dynamics of a chimeric variant of the SH3 domain (SHA-Bergerac) by NMR spectroscopy
Russ.J.Bioorganic Chem., 34, 2008
|
|
2RQW
 
 | |
2ROT
 
 | | Structure of chimeric variant of SH3 domain- SHH | | Descriptor: | Spectrin alpha chain, brain | | Authors: | Kutyshenko, N.P, Prokhorov, D.A, Timchenko, M.A, Kudrevatykh, Y.A, Gushchina, L.V, Khristoforov, V.S, Filimonov, V.V. | | Deposit date: | 2008-04-10 | | Release date: | 2009-04-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the chimeric SH3 domains, SHH- and SHA-"Bergeracs".
Biochim.Biophys.Acta, 1794, 2009
|
|
2RPN
 
 | | A crucial role for high intrinsic specificity in the function of yeast SH3 domains | | Descriptor: | Actin-binding protein, Actin-regulating kinase 1 | | Authors: | Stollar, E.J, Garcia, B, Chong, A, Forman-Kay, J, Davidson, A. | | Deposit date: | 2008-06-12 | | Release date: | 2009-06-16 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural, functional, and bioinformatic studies demonstrate the crucial role of an extended peptide binding site for the SH3 domain of yeast Abp1p
J.Biol.Chem., 284, 2009
|
|
2SEM
 
 | | SEM5 SH3 DOMAIN COMPLEXED WITH PEPTOID INHIBITOR | | Descriptor: | PROTEIN (SEX MUSCLE ABNORMAL PROTEIN 5), PROTEIN (SH3 PEPTOID INHIBITOR) | | Authors: | Nguyen, J.T, Turck, C.W, Cohen, F.E, Zuckermann, R.N, Lim, W.A. | | Deposit date: | 1998-11-02 | | Release date: | 1999-01-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploiting the basis of proline recognition by SH3 and WW domains: design of N-substituted inhibitors.
Science, 282, 1998
|
|
2RN8
 
 | |
2V1R
 
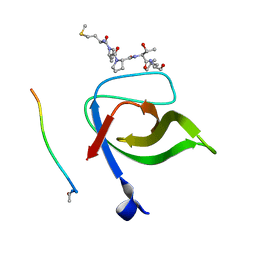 | | Yeast Pex13 SH3 domain complexed with a peptide from Pex14 at 2.1 A resolution | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PAS20, PEX14 | | Authors: | Kursula, I, Kursula, P, Lehmann, F, Zou, P, Song, Y.H, Wilmanns, M. | | Deposit date: | 2007-05-29 | | Release date: | 2008-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Genomics of Yeast SH3 Domains
To be Published
|
|
2VKN
 
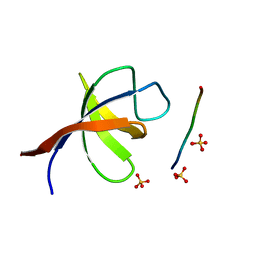 | | YEAST SHO1 SH3 DOMAIN COMPLEXED WITH A PEPTIDE FROM PBS2 | | Descriptor: | MAP KINASE KINASE PBS2, PROTEIN SSU81, SULFATE ION | | Authors: | Kursula, P, Kursula, I, Song, Y.H, Paraskevopoulos, K, Wilmanns, M. | | Deposit date: | 2007-12-20 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Genomics of Yeast SH3 Domains
To be Published
|
|
2VWF
 
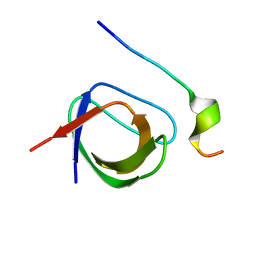 | | Grb2 SH3C (2) | | Descriptor: | GRB2-ASSOCIATED-BINDING PROTEIN 2, GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2 | | Authors: | Harkiolaki, M, Tsirka, T, Feller, S.M. | | Deposit date: | 2008-06-24 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Distinct Binding Modes of Two Epitopes in Gab2 that Interact with the Sh3C Domain of Grb2.
Structure, 17, 2009
|
|
2V1Q
 
 | | Atomic-resolution structure of the yeast Sla1 SH3 domain 3 | | Descriptor: | CHLORIDE ION, CYTOSKELETON ASSEMBLY CONTROL PROTEIN SLA1, PLATINUM (II) ION, ... | | Authors: | Kursula, I, Kursula, P, Zou, P, Lehmann, F, Song, Y.H, Wilmanns, M. | | Deposit date: | 2007-05-29 | | Release date: | 2008-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Genomics of Yeast SH3 Domains
To be Published
|
|
2W10
 
 | | Mona SH3C in complex | | Descriptor: | GRB2-RELATED ADAPTOR PROTEIN 2, PHOSPHATE ION, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 23 | | Authors: | Harkiolaki, M, Feller, S.M. | | Deposit date: | 2008-10-13 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct Binding Modes of Two Epitopes in Gab2 that Interact with the Sh3C Domain of Grb2.
Structure, 17, 2009
|
|
2VVK
 
 | | Grb2 SH3C (1) | | Descriptor: | GLYCEROL, GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2 | | Authors: | Harkiolaki, M, Tsirka, T, Feller, S.M. | | Deposit date: | 2008-06-09 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Distinct Binding Modes of Two Epitopes in Gab2 that Interact with the Sh3C Domain of Grb2.
Structure, 17, 2009
|
|
2W0Z
 
 | | Grb2 SH3C (3) | | Descriptor: | GRB2-ASSOCIATED-BINDING PROTEIN 2, GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2 | | Authors: | Harkiolaki, M, Tsirka, T, Feller, S.M. | | Deposit date: | 2008-10-13 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Distinct Binding Modes of Two Epitopes in Gab2 that Interact with the Sh3C Domain of Grb2.
Structure, 17, 2009
|
|
5OB1
 
 | |
5OB0
 
 | |
5NP2
 
 | | Abl1 SH3 pTyr89/134 | | Descriptor: | Tyrosine-protein kinase ABL1 | | Authors: | Mero, B, Radnai, L, Gogl, G, Leveles, I, Buday, L. | | Deposit date: | 2017-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the tyrosine phosphorylation-mediated inhibition of SH3 domain-ligand interactions.
J.Biol.Chem., 294, 2019
|
|
5O2P
 
 | | p130Cas SH3 domain PTP-PEST peptide chimera | | Descriptor: | Breast cancer anti-estrogen resistance 1,Tyrosine-protein phosphatase non-receptor type 12 | | Authors: | Hexnerova, R, Veverka, V. | | Deposit date: | 2017-05-22 | | Release date: | 2017-09-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of CAS SH3 domain selectivity and regulation reveals new CAS interaction partners.
Sci Rep, 7, 2017
|
|
