2CCW
 
 | | Crystal structure of Azurin II at atomic resolution (1.13 angstrom) | | Descriptor: | AZURIN II, COPPER (I) ION | | Authors: | Paraskevopoulos, K, Hough, M.A, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2006-01-18 | | Release date: | 2006-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Active Site Structures and the Redox Properties of Blue Copper Proteins: Atomic Resolution Structure of Azurin II and Electronic Structure Calculations of Azurin, Plastocyanin and Stellacyanin.
Dalton Trans., 25, 2006
|
|
2IWK
 
 | | Inhibitor-bound form of nitrous oxide reductase from Achromobacter Cycloclastes at 1.7 Angstrom resolution | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Paraskevopoulos, K, Antonyuk, S.V, Sawers, R.G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2006-06-30 | | Release date: | 2006-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insight Into Catalysis of Nitrous Oxide Reductase from High-Resolution Structures of Resting and Inhibitor-Bound Enzyme from Achromobacter Cycloclastes.
J.Mol.Biol., 362, 2006
|
|
2IWF
 
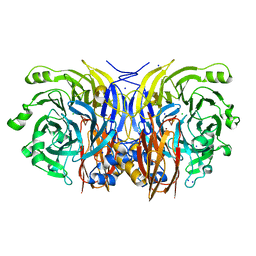 | | Resting form of pink nitrous oxide reductase from Achromobacter Cycloclastes | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Paraskevopoulos, K, Antonyuk, S.V, Sawers, R.G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2006-06-29 | | Release date: | 2006-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Insight Into Catalysis of Nitrous Oxide Reductase from High-Resolution Structures of Resting and Inhibitor-Bound Enzyme from Achromobacter Cycloclastes.
J.Mol.Biol., 362, 2006
|
|
2JFC
 
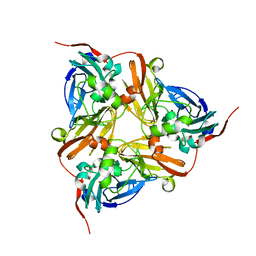 | | M144L mutant of Nitrite Reductase from Alcaligenes xylosoxidans in space group P212121 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE | | Authors: | Paraskevopoulos, K, Hough, M.A, Sawers, R.G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2007-01-31 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of the met144Leu Mutant of Copper Nitrite Reductase from Alcaligenes Xylosoxidans Provides the First Glimpse of a Protein-Protein Complex with Azurin II.
J.Biol.Inorg.Chem., 12, 2007
|
|
4B0Z
 
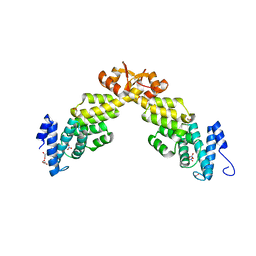 | | Crystal structure of S. pombe Rpn12 | | Descriptor: | 26S PROTEASOME REGULATORY SUBUNIT RPN12, GLYCEROL, MONOTHIOGLYCEROL, ... | | Authors: | Boehringer, J, Riedinger, C, Paraskevopoulos, K, Johnson, E.O.D, Lowe, E.D, Khoudian, C, Smith, D, Noble, M.E.M, Gordon, C, Endicott, J.A. | | Deposit date: | 2012-07-06 | | Release date: | 2012-09-12 | | Last modified: | 2012-11-07 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | Structural and Functional Characterisation of Rpn12 Identifies Residues Required for Rpn10 Proteasome Incorporation.
Biochem.J., 448, 2012
|
|
2VKN
 
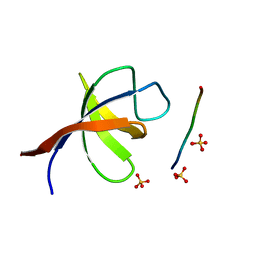 | | YEAST SHO1 SH3 DOMAIN COMPLEXED WITH A PEPTIDE FROM PBS2 | | Descriptor: | MAP KINASE KINASE PBS2, PROTEIN SSU81, SULFATE ION | | Authors: | Kursula, P, Kursula, I, Song, Y.H, Paraskevopoulos, K, Wilmanns, M. | | Deposit date: | 2007-12-20 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Genomics of Yeast SH3 Domains
To be Published
|
|
