6WXP
 
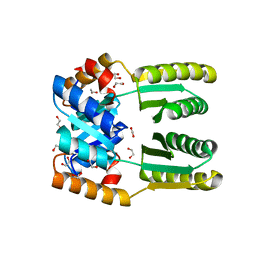 | | De novo TIM barrel-ferredoxin fold fusion homodimer with 4-glutamate centre TFD-EE | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, TFD-EE | | Authors: | Caldwell, S.J, Zeymer, C, Haydon, I.C, Huang, P, Hilvert, D, Baker, D. | | Deposit date: | 2020-05-11 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tight and specific lanthanide binding in a de novo TIM barrel with a large internal cavity designed by symmetric domain fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5C2M
 
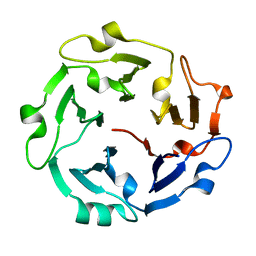 | | The de novo evolutionary emergence of a symmetrical protein is shaped by folding constraints | | Descriptor: | Predicted protein | | Authors: | Smock, R.G, Yadid, I, Dym, O, Clarke, J, Tawfik, D.S. | | Deposit date: | 2015-06-16 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De Novo Evolutionary Emergence of a Symmetrical Protein Is Shaped by Folding Constraints.
Cell, 164, 2016
|
|
5C2N
 
 | | The de novo evolutionary emergence of a symmetrical protein is shaped by folding constraints | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta propeller | | Authors: | Smock, R.G, Yadid, I, Dym, O, Clarke, J, Tawfik, D.S. | | Deposit date: | 2015-06-16 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | De Novo Evolutionary Emergence of a Symmetrical Protein Is Shaped by Folding Constraints.
Cell, 164, 2016
|
|
7Q1R
 
 | | A de novo designed homo-dimeric antiparallel coiled coil apCC-Di | | Descriptor: | ETHANOL, SODIUM ION, apCC-Di | | Authors: | Shanmugaratnam, S, Rhys, G.G, Dawson, W.M, Woolfson, D.N, Hocker, B. | | Deposit date: | 2021-10-20 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | De novo designed peptides for cellular delivery and subcellular localisation.
Nat.Chem.Biol., 18, 2022
|
|
7Q1S
 
 | | A de novo designed hetero-dimeric antiparallel coiled coil apCC-Di-AB_var | | Descriptor: | 1,2-ETHANEDIOL, apCC-Di-A_var, apCC-Di-B_var | | Authors: | Shanmugaratnam, S, Rhys, G.G, Dawson, W.M, Woolfson, D.N, Hocker, B. | | Deposit date: | 2021-10-20 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | De novo designed peptides for cellular delivery and subcellular localisation.
Nat.Chem.Biol., 18, 2022
|
|
7Q1T
 
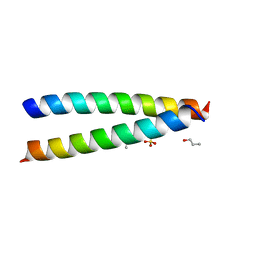 | | A de novo designed hetero-dimeric antiparallel coiled coil apCC-Di-AB | | Descriptor: | N-PROPANOL, SULFATE ION, apCC-Di-A, ... | | Authors: | Shanmugaratnam, S, Rhys, G.G, Dawson, W.M, Woolfson, D.N, Hocker, B. | | Deposit date: | 2021-10-20 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | De novo designed peptides for cellular delivery and subcellular localisation.
Nat.Chem.Biol., 18, 2022
|
|
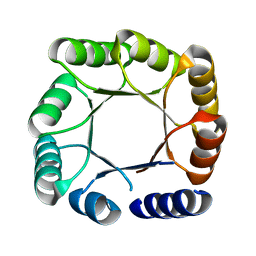
5BVL
 
 | |
8URE
 
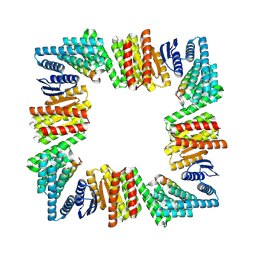 | |
7UEK
 
 | |
8VL4
 
 | |
8VL3
 
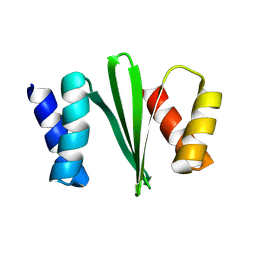 | |
5E6G
 
 | |
6W90
 
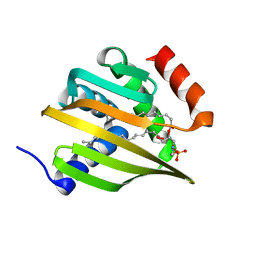 | | De novo designed NTF2 fold protein NT-9 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, NTF2 fold protein loop-helix-loop design NT-9 | | Authors: | Thompson, M.C, Pan, X, Liu, L, Fraser, J.S, Kortemme, T. | | Deposit date: | 2020-03-21 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Expanding the space of protein geometries by computational design of de novo fold families.
Science, 369, 2020
|
|
1LQ7
 
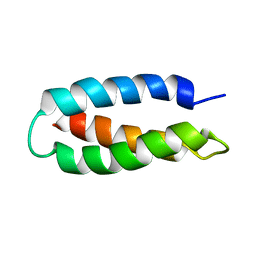 | | De Novo Designed Protein Model of Radical Enzymes | | Descriptor: | Alpha3W | | Authors: | Dai, Q.-H, Tommos, C, Fuentes, E.J, Blomberg, M, Dutton, P.L, Wand, A.J. | | Deposit date: | 2002-05-09 | | Release date: | 2002-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a De Novo Designed Protein Model of Radical Enzymes
J.Am.Chem.Soc., 124, 2002
|
|
8BL3
 
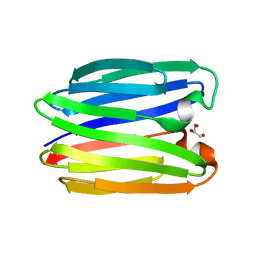 | |
8BL6
 
 | |
7DGW
 
 | | De novo designed protein H4A2S | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, de novo designed protein H4A2S | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
4KY3
 
 | | Three-dimensional Structure of the orthorhombic crystal of computationally designed insertion domain , Northeast Structural Genomics Consortium (NESG) Target OR327 | | Descriptor: | designed protein OR327 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Gleixner, J, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-28 | | Release date: | 2013-06-19 | | Last modified: | 2016-02-17 | | Method: | X-RAY DIFFRACTION (2.964 Å) | | Cite: | Precise assembly of complex beta sheet topologies from de novo designed building blocks.
Elife, 4, 2015
|
|
6XEH
 
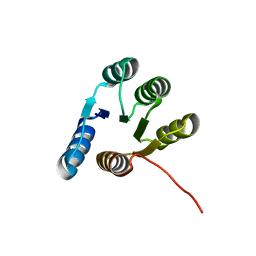 | |
6X8N
 
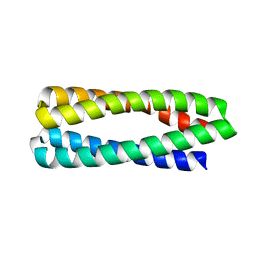 | | Crystal Structure of H49A ABLE mutant | | Descriptor: | De novo designed ABLE protein | | Authors: | Polizzi, N.F. | | Deposit date: | 2020-06-01 | | Release date: | 2020-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A defined structural unit enables de novo design of small-molecule-binding proteins.
Science, 369, 2020
|
|
7TJL
 
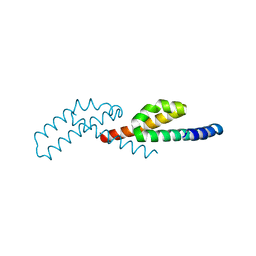 | |
2MTL
 
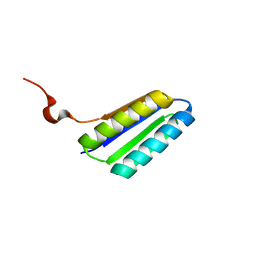 | | Solution NMR Structure of De novo designed FR55, Northeast Structural Genomics Consortium (NESG) Target OR109 | | Descriptor: | De novo designed protein FR55 OR109 | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Hamilton, K, Ciccosanti, C, Sahdev, S, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed FR55, Northeast Structural Genomics Consortium (NESG) Target OR109
To be Published
|
|
2N75
 
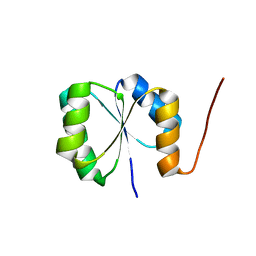 | | Solution NMR Structure of De novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446 | | Descriptor: | De novo designed protein | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446
To be Published
|
|
8AH9
 
 | |
8R8O
 
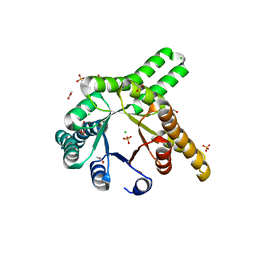 | | Hallucinated de novo TIM barrel with three helical extensions - HalluTIM3-1 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Beck, J, Shanmugaratnam, S, Hocker, B. | | Deposit date: | 2023-11-29 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Diversifying de novo TIM barrels by hallucination.
Protein Sci., 33, 2024
|
|
