3QLH
 
 | | HIV-1 Reverse Transcriptase in Complex with Manicol at the RNase H Active Site and TMC278 (Rilpivirine) at the NNRTI Binding Pocket | | Descriptor: | (2S)-5,7-dihydroxy-9-methyl-2-(prop-1-en-2-yl)-1,2,3,4-tetrahydro-6H-benzo[7]annulen-6-one, 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Himmel, D.M, Wojtak, K, Bauman, J.D, Arnold, E. | | Deposit date: | 2011-02-02 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthesis, activity, and structural analysis of novel alpha-hydroxytropolone inhibitors of human immunodeficiency virus reverse transcriptase-associated ribonuclease H.
J.Med.Chem., 54, 2011
|
|
3G43
 
 | |
3G4Z
 
 | | Crystal Structure of NiSOD Y9F mutant at 1.9 A | | Descriptor: | BROMIDE ION, NICKEL (II) ION, Superoxide dismutase [Ni] | | Authors: | Garman, S.C, Guce, A.I, Herbst, R.W, Bryngelson, P.A, Cabelli, D.E, Higgins, K.A, Ryan, K.C, Maroney, M.J. | | Deposit date: | 2009-02-04 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Role of conserved tyrosine residues in NiSOD catalysis: a case of convergent evolution
Biochemistry, 48, 2009
|
|
3G5D
 
 | | Kinase domain of cSrc in complex with Dasatinib | | Descriptor: | GLYCEROL, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Grutter, C, Kluter, S, Rauh, D. | | Deposit date: | 2009-02-05 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hybrid compound design to overcome the gatekeeper T338M mutation in cSrc
J.Med.Chem., 52, 2009
|
|
3FUR
 
 | | Crystal Structure of PPARg in complex with INT131 | | Descriptor: | 2,4-dichloro-N-[3,5-dichloro-4-(quinolin-3-yloxy)phenyl]benzenesulfonamide, CHLORIDE ION, Nuclear receptor coactivator 1, ... | | Authors: | Wang, Z, Liu, J, Walker, N. | | Deposit date: | 2009-01-14 | | Release date: | 2009-06-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | INT131: a selective modulator of PPAR gamma.
J.Mol.Biol., 386, 2009
|
|
3QGJ
 
 | | 1.3A Structure of alpha-Lytic Protease Bound to Ac-AlaAlaPro-Alanal | | Descriptor: | 1,2-ETHANEDIOL, Ac-AlaAlaPro-Alanal peptide, Alpha-lytic protease, ... | | Authors: | Everill, P, Meinke, G, Bohm, A, Bachovchin, W. | | Deposit date: | 2011-01-24 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Substrate Binding Defines Ser195 Position in the Catalytic Triad of Serine Proteases, Not His57 protonation
To be Published
|
|
3G84
 
 | |
3QO1
 
 | | Monoclinic form of IgG1 Fab fragment (apo form) sharing same Fv as IgA | | Descriptor: | Fab fragment of IMMUNOGLOBULIN G1 HEAVY CHAIN, Fab fragment of IMMUNOGLOBULIN G1 LIGHT CHAIN, GLYCEROL | | Authors: | Trajtenberg, F, Correa, A, Buschiazzo, A. | | Deposit date: | 2011-02-09 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a human IgA1 Fab fragment at 1.55 angstrom resolution: potential effect of the constant domains on antigen-affinity modulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3QPN
 
 | | Structure of PDE10-inhibitor complex | | Descriptor: | 6-methoxy-7-[2-(quinolin-2-yl)ethoxy]quinazoline, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Pandit, J, Marr, E.S. | | Deposit date: | 2011-02-14 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Use of Structure-Based Design to Discover a Potent, Selective, In Vivo Active Phosphodiesterase 10A Inhibitor Lead Series for the Treatment of Schizophrenia.
J.Med.Chem., 54, 2011
|
|
3QH7
 
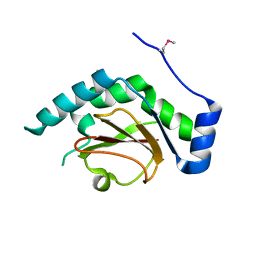 | | 2.5 A resolution structure of Se-Met labeled CT296 from Chlamydia trachomatis | | Descriptor: | CT296 | | Authors: | Kemege, K, Hickey, J, Lovell, S, Battaile, K.P, Zhang, Y, Hefty, P.S. | | Deposit date: | 2011-01-25 | | Release date: | 2011-10-05 | | Last modified: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Ab initio structural modeling of and experimental validation for Chlamydia trachomatis protein CT296 reveal structural similarity to Fe(II) 2-oxoglutarate-dependent enzymes.
J. Bacteriol., 193, 2011
|
|
3FVQ
 
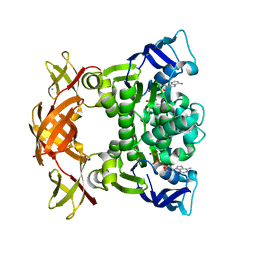 | | Crystal structure of the nucleotide binding domain FbpC complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Fe(3+) ions import ATP-binding protein fbpC | | Authors: | Newstead, S, Bilton, P, Carpenter, E.P, Campopiano, D, Iwata, S. | | Deposit date: | 2009-01-16 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into how nucleotide-binding domains power ABC transport.
Structure, 17, 2009
|
|
3QHX
 
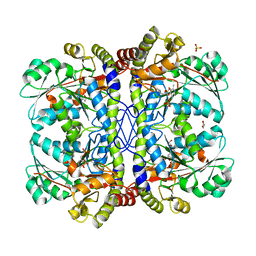 | |
3QR9
 
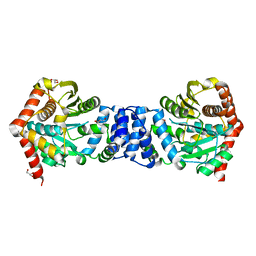 | |
3FX0
 
 | |
3QIX
 
 | | Crystal Structure of BoNT/A LC with Zinc bound | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin type A, ZINC ION | | Authors: | Thompson, A.A, Han, G.W, Stevens, R.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structural Characterization of Three Novel Hydroxamate-Based Zinc Chelating Inhibitors of the Clostridium botulinum Serotype A Neurotoxin Light Chain Metalloprotease Reveals a Compact Binding Site Resulting from 60/70 Loop Flexibility.
Biochemistry, 50, 2011
|
|
3GAH
 
 | | Structure of a F112H variant PduO-type ATP:corrinoid adenosyltransferase from Lactobacillus reuteri complexed with cobalamin and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COBALAMIN, Cobalamin adenosyltransferase PduO-like protein, ... | | Authors: | St Maurice, M, Mera, P.E, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2009-02-17 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Residue Phe112 of the human-type corrinoid adenosyltransferase (PduO) enzyme of Lactobacillus reuteri is critical to the formation of the four-coordinate Co(II) corrinoid substrate and to the activity of the enzyme.
Biochemistry, 48, 2009
|
|
3QSI
 
 | |
3QU1
 
 | | Peptide deformylase from Vibrio cholerae | | Descriptor: | CHLORIDE ION, Peptide deformylase 2, SULFATE ION, ... | | Authors: | Osipiuk, J, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-23 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptide deformylase from Vibrio cholerae.
To be Published
|
|
3QK7
 
 | | Crystal structure of putative Transcriptional regulator from Yersinia pestis biovar Microtus str. 91001 | | Descriptor: | Transcriptional regulators | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of putative Transcriptional regulator from Yersinia pestis biovar Microtus str. 91001
To be Published
|
|
3GCN
 
 | |
3QUF
 
 | | The structure of a family 1 extracellular solute-binding protein from Bifidobacterium longum subsp. infantis | | Descriptor: | ACETIC ACID, Extracellular solute-binding protein, family 1, ... | | Authors: | Cuff, M.E, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-23 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a family 1 extracellular solute-binding protein from Bifidobacterium longum subsp. infantis
TO BE PUBLISHED
|
|
3FYM
 
 | | The 1A structure of YmfM, a putative DNA-binding membrane protein from Staphylococcus aureus | | Descriptor: | Putative uncharacterized protein, ZINC ION | | Authors: | Xu, L, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2009-01-22 | | Release date: | 2010-02-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The 1A structure of YmfM, a putative DNA-binding membrane protein from Staphylococcus aureus
To be Published
|
|
3QL2
 
 | |
3QV2
 
 | |
3FZI
 
 | |
