3FW9
 
 | | Structure of berberine bridge enzyme in complex with (S)-scoulerine | | Descriptor: | (13aS)-3,10-dimethoxy-5,8,13,13a-tetrahydro-6H-isoquino[3,2-a]isoquinoline-2,9-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Winkler, A, Macheroux, P, Gruber, K. | | Deposit date: | 2009-01-17 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Structural roles of biocovalent flaninylation in berberine bridge enzyme
to be published
|
|
3QST
 
 | |
3QAH
 
 | | Crystal structure of apo-form human MOF catalytic domain | | Descriptor: | Probable histone acetyltransferase MYST1, ZINC ION | | Authors: | Sun, B, Tang, Q, Zhong, C, Ding, J. | | Deposit date: | 2011-01-11 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Regulation of the histone acetyltransferase activity of hMOF via autoacetylation of Lys274
Cell Res., 21, 2011
|
|
3QAW
 
 | |
3FXM
 
 | | Crystal Structure of Human Protein phosphatase 1A (PPM1A) Bound with Citrate at 10 mM of Mn2+ | | Descriptor: | CITRATE ANION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Hu, T, Wang, L, Wang, K, Jiang, H, Shen, X. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the Mn2+-dependent activation of human PPM1A
To be published
|
|
3FWE
 
 | | Crystal Structure of the Apo D138L CAP mutant | | Descriptor: | Catabolite gene activator, PROLINE | | Authors: | Sharma, H, Wang, J, Kong, J, Yu, S, Steitz, T. | | Deposit date: | 2009-01-17 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of apo-CAP reveals that large conformational changes are necessary for DNA binding
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3FXX
 
 | | Human EphA3 Kinase and Juxtamembrane Region Bound to Substrate KQWDNYE[pTyr]IW | | Descriptor: | Ephrin type-A receptor 3, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Davis, T, Walker, J.R, Mackenzie, F, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-21 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural recognition of an optimized substrate for the ephrin family of receptor tyrosine kinases.
Febs J., 276, 2009
|
|
3FYA
 
 | | Crystal Structure of an R35A mutant of the Restriction-Modification Controller Protein C.Esp1396I | | Descriptor: | Regulatory protein | | Authors: | Ball, N.J, McGeehan, J.E, Thresh, S.J, Streeter, S.D, Kneale, G.G. | | Deposit date: | 2009-01-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the restriction-modification controller protein C.Esp1396I.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3QEH
 
 | | Crystal structure of human N12-i15, an ADCC and non-neutralizing anti-HIV-1 Env antibody | | Descriptor: | CHLORIDE ION, Fab fragment of human anti-HIV-1 Env antibody N12-i15, heavy chain, ... | | Authors: | Guan, Y, DeVico, A.L, Lewis, G.K, Pazgier, M. | | Deposit date: | 2011-01-20 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of human N12-i15, an ADCC and non-neutralizing anti-HIV-1 Env antibody
To be Published
|
|
3QCB
 
 | | Human receptor protein tyrosine phosphatase gamma, domain 1, apo | | Descriptor: | Receptor-type tyrosine-protein phosphatase gamma, SULFATE ION | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-16 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small molecule receptor protein tyrosine phosphatase [gamma](RPTP[gamma]) ligands that inhibit phosphatase activity via perturbation of the tryptophan-proline-aspartate (WPD) loop
J.Med.Chem., 54, 2011
|
|
3FYN
 
 | | Crystal structure from the mobile metagenome of Cole Harbour Salt Marsh: Integron Cassette Protein HFX_CASS3 | | Descriptor: | ACETATE ION, Integron gene cassette protein HFX_CASS3, MAGNESIUM ION | | Authors: | Sureshan, V, Deshpande, C.N, Harrop, S.J, Kudritska, M, Koenig, J.E, Evdokimova, E, Osipiuk, J, Edwards, A.M, Savchenko, A, Joachimiak, A, Doolittle, W.F, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-22 | | Release date: | 2009-02-10 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structure from the mobile metagenome of Cole Harbour Salt Marsh: Integron Cassette Protein HFX_CASS3
To be Published
|
|
3QFD
 
 | |
3QGP
 
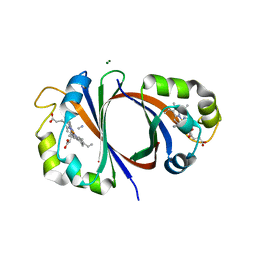 | |
3G0K
 
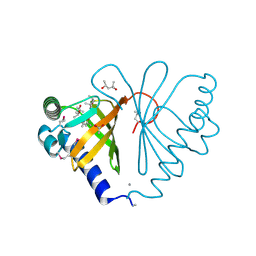 | |
3QD0
 
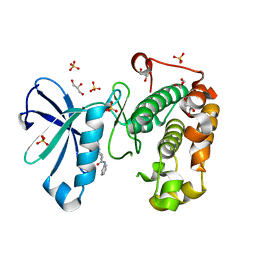 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with (2R,5S)-1-[2-Amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-6-methyl-N-phenyl-3-piperidinecarboxamide | | Descriptor: | (3S,6R)-1-[2-amino-6-(3-amino-2H-indazol-6-yl)pyrimidin-4-yl]-6-methyl-N-phenylpiperidine-3-carboxamide, 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3G0Z
 
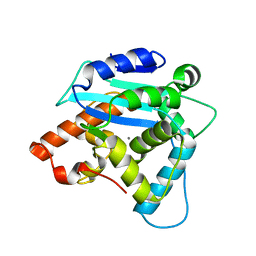 | | Structure of S. pombe Pop2p - Zn2+ and Mn2+ bound form | | Descriptor: | CCR4-Not complex subunit Caf1, MANGANESE (II) ION, ZINC ION | | Authors: | Andersen, K.R, Jonstrup, A.T, Van, L.B, Brodersen, D.E. | | Deposit date: | 2009-01-29 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | The activity and selectivity of fission yeast Pop2p are affected by a high affinity for Zn2+ and Mn2+ in the active site
Rna, 15, 2009
|
|
3QDL
 
 | | Crystal structure of RdxA from Helicobacter pyroli | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Oxygen-insensitive NADPH nitroreductase | | Authors: | Rojas, A.L, Martinez-Julvez, M, Olekhnovich, I.N, Hoffman, P.S, Sancho, J. | | Deposit date: | 2011-01-18 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of RdxA--an oxygen-insensitive nitroreductase essential for metronidazole activation in Helicobacter pylori.
Febs J., 279, 2012
|
|
3QID
 
 | | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | | Descriptor: | GLYCEROL, MANGANESE (III) ION, RNA dependent RNA polymerase, ... | | Authors: | Kim, K.H, Intekhab, A, Lee, J.H. | | Deposit date: | 2011-01-27 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase.
J.Gen.Virol., 92, 2011
|
|
3GI6
 
 | | Crystal structure of protease inhibitor, AD78 in complex with wild type HIV-1 protease | | Descriptor: | (5S)-N-[(1S,2R)-2-Hydroxy-3-[[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino]-1-(phenylmethyl)propyl]-2-oxo-3-[3-(trif luoromethyl)phenyl]-5-oxazolidinecarboxamide, PHOSPHATE ION, Protease | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2009-03-05 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Evaluating the substrate-envelope hypothesis: structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance.
J.Virol., 84, 2010
|
|
3G2S
 
 | | VHS Domain of human GGA1 complexed with SorLA C-terminal Peptide | | Descriptor: | ADP-ribosylation factor-binding protein GGA1, C-terminal fragment of Sortilin-related receptor, HEXANE-1,6-DIOL | | Authors: | Cramer, J.F, Behrens, M.A, Gustafsen, C, Oliveira, C.L.P, Pedersen, J.S, Madsen, P, Petersen, C.M, Thirup, S.S. | | Deposit date: | 2009-02-01 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | GGA autoinhibition revisited
Traffic, 11, 2010
|
|
3QJZ
 
 | | Crystal structure of PI3K-gamma in complex with benzothiazole 1 | | Descriptor: | N-{6-[2-(methylsulfanyl)pyrimidin-4-yl]-1,3-benzothiazol-2-yl}acetamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors.
J.Med.Chem., 54, 2011
|
|
3G38
 
 | | The catalytically inactive mutant Mth0212 (D151N) in complex with an 8 bp dsDNA | | Descriptor: | 5'-D(*CP*CP*TP*GP*UP*GP*CP*GP*AP*T)-3', 5'-D(*CP*GP*CP*GP*CP*AP*GP*GP*C)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-02 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3GJ1
 
 | | Non photoactivated state of PA-GFP | | Descriptor: | CHLORIDE ION, Green fluorescent protein, SULFATE ION | | Authors: | Henderson, J.N, Gepshtein, R, Heenan, J.R, Kallio, K, Huppert, D, Remington, S.J. | | Deposit date: | 2009-03-07 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of the photoactivatable green fluorescent protein.
J.Am.Chem.Soc., 131, 2009
|
|
3QL9
 
 | |
3FT7
 
 | | Crystal structure of an extremely stable dimeric protein from sulfolobus islandicus | | Descriptor: | GLYCEROL, Uncharacterized protein ORF56 | | Authors: | Neumann, P, Loew, C, Weininger, U, Stubbs, M.T. | | Deposit date: | 2009-01-12 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Stability Analysis of an Extremely Stable Dimeric DNA Binding Protein from Sulfolobus islandicus
Biochemistry, 48, 2009
|
|
