3NY4
 
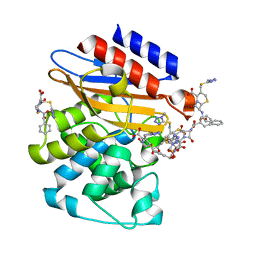 | | Crystal Structure of BlaC-K73A bound with Cefamandole | | Descriptor: | (6R,7R)-7-{[(2R)-2-hydroxy-2-phenylacetyl]amino}-3-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Tremblay, L.W, Blanchard, J.S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structures of the Michaelis Complex (1.2 A) and the Covalent Acyl Intermediate (2.0 A) of Cefamandole Bound in the Active Sites of the Mycobacterium tuberculosis beta-Lactamase K73A and E166A Mutants.
Biochemistry, 49, 2010
|
|
3NYY
 
 | |
3O75
 
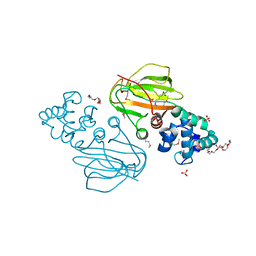
 | | Crystal structure of Cra transcriptional dual regulator from Pseudomonas putida in complex with fructose 1-phosphate' | | Descriptor: | 1-O-phosphono-beta-D-fructofuranose, Fructose transport system repressor FruR | | Authors: | Chavarria, M, Santiago, C, Platero, R, Krell, T, Casasnovas, J.M, de Lorenzo, V. | | Deposit date: | 2010-07-30 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fructose 1-phosphate is the preferred effector of the metabolic regulator Cra of Pseudomonas putida
J.Biol.Chem., 286, 2011
|
|
3O13
 
 | |
3O2B
 
 | | E. coli ClpS in complex with a Phe N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS, CHLORIDE ION, Phe N-end rule peptide, ... | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3O89
 
 | | Crystal Structure of Sperm Whale Myoglobin G65T | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Huang, X, Lovelace, L, Lebioda, L. | | Deposit date: | 2010-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Investigations of Structural Factors that Influence the Mechanism of Halophenol Dehalogenation using 'Peroxidase-Like' Myoglobin mutants and 'Myoglobin-Like' Amphitrite ornata Dehaloperoxidase Mutants
TO BE PUBLISHED
|
|
3O8I
 
 | | Structure of 14-3-3 isoform sigma in complex with a C-Raf1 peptide and a stabilizing small molecule fragment | | Descriptor: | 14-3-3 binding site peptide of RAF proto-oncogene serine/threonine-protein kinase, 14-3-3 protein sigma, 6,6-dihydroxy-1-methoxyhexan-2-one | | Authors: | Ottmann, C, Rose, R, Kaiser, M, Kuhenne, P. | | Deposit date: | 2010-08-03 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Impaired Binding of 14-3-3 to C-RAF in Noonan Syndrome Suggests New Approaches in Diseases with Increased Ras Signaling
Mol.Cell.Biol., 30, 2010
|
|
3O3A
 
 | | Human Class I MHC HLA-A2 in complex with the Peptidomimetic ELA-1 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Borbulevych, O.Y, Baker, B.M. | | Deposit date: | 2010-07-23 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of HLA-A*0201 complexed with Melan-A/MART-1(26(27L)-35) peptidomimetics reveal conformational heterogeneity and highlight degeneracy of T cell recognition.
J.Med.Chem., 53, 2010
|
|
3O4V
 
 | |
3O9H
 
 | | Crystal Structure of wild-type HIV-1 Protease in complex with kd26 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(1S,2R)-3-[(1,3-benzodioxol-5-ylsulfonyl)(2-ethylbutyl)amino]-1-benzyl-2-hydroxypropyl}carbamate, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
3OA6
 
 | | Human MSL3 Chromodomain bound to DNA and H4K20me1 peptide | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*A)-3'), H4 peptide monomethylated at lysine 20, ... | | Authors: | Kim, D, Huang, P, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2010-08-04 | | Release date: | 2010-08-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Corecognition of DNA and a methylated histone tail by the MSL3 chromodomain.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3O7A
 
 | | Crystal structure of PHF13 in complex with H3K4me3 | | Descriptor: | GLYCEROL, H3K4ME3 HISTONE 11MER-PEPTIDE, PHD finger protein 13 variant, ... | | Authors: | Bian, C.B, Lam, R, Xu, C, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-30 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | PHF13 is a molecular reader and transcriptional co-regulator of H3K4me2/3.
Elife, 5, 2016
|
|
3OAP
 
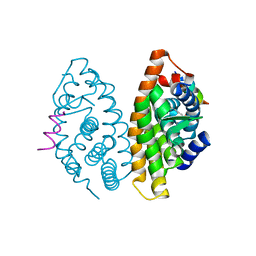 | | Crystal structure of human Retinoid X Receptor alpha-ligand binding domain complex with 9-cis retinoic acid and the coactivator peptide GRIP-1 | | Descriptor: | (9cis)-retinoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Smith, C.D, Muccio, D.D. | | Deposit date: | 2010-08-05 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure, Energetics and Dynamics of Binding Coactivator Peptide to Human Retinoid X Receptor Alpha Ligand Binding Domain Complex with 9-cis-Retinoic Acid.
Biochemistry, 50, 2011
|
|
3OB1
 
 | |
3OBE
 
 | |
3OAK
 
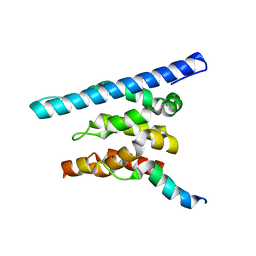 | | Crystal structure of a Spn1 (Iws1)-Spt6 complex | | Descriptor: | Transcription elongation factor SPT6, Transcription factor IWS1 | | Authors: | McDonald, S.M, Close, D, Hill, C.P. | | Deposit date: | 2010-08-05 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and biological importance of the spn1-spt6 interaction, and its regulatory role in nucleosome binding.
Mol.Cell, 40, 2010
|
|
3NOM
 
 | | Crystal Structure of Zymomonas mobilis Glutaminyl Cyclase (monoclinic form) | | Descriptor: | CALCIUM ION, GLYCEROL, Glutamine cyclotransferase, ... | | Authors: | Parthier, C, Carrillo, D.R, Stubbs, M.T. | | Deposit date: | 2010-06-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic and structural characterization of bacterial glutaminyl cyclases from Zymomonas mobilis and Myxococcus xanthus
Biol.Chem., 391, 2010
|
|
3NS9
 
 | | Crystal structure of CDK2 in complex with inhibitor BS-194 | | Descriptor: | (2S,3S)-3-{[7-(benzylamino)-3-(1-methylethyl)pyrazolo[1,5-a]pyrimidin-5-yl]amino}butane-1,2,4-triol, Cell division protein kinase 2 | | Authors: | Hazel, P, Freemont, P.S. | | Deposit date: | 2010-07-01 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Novel Pyrazolo[1,5-a]pyrimidine Is a Potent Inhibitor of Cyclin-Dependent Protein Kinases 1, 2, and 9, Which Demonstrates Antitumor Effects in Human Tumor Xenografts Following Oral Administration.
J.Med.Chem., 53, 2010
|
|
3NT3
 
 | | CRYSTAL STRUCTURE OF LSSmKate2 red fluorescent proteins with large Stokes shift | | Descriptor: | GLYCEROL, LSSmKate2 red fluorescent protein | | Authors: | Malashkevich, V.N, Piatkevich, K, Almo, S.C, Verkhusha, V. | | Deposit date: | 2010-07-02 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering ESPT Pathways Based on Structural Analysis of LSSmKate Red Fluorescent Proteins with Large Stokes Shift.
J.Am.Chem.Soc., 132, 2010
|
|
3NTR
 
 | | Crystal structure of K97V mutant of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NAD and inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6503 Å) | | Cite: | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
3NRG
 
 | |
3NSG
 
 | | Crystal Structure of OmpF, an Outer Membrane Protein from Salmonella typhi | | Descriptor: | CITRATE ANION, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Balasubramaniam, D, Arockiasamy, A, Sharma, A, Krishnaswamy, S. | | Deposit date: | 2010-07-01 | | Release date: | 2011-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Asymmetric pore occupancy in crystal structure of OmpF porin from Salmonella typhi
J.Struct.Biol., 178, 2012
|
|
3NUZ
 
 | |
3NWP
 
 | |
3OD8
 
 | | Human PARP-1 zinc finger 1 (Zn1) bound to DNA | | Descriptor: | 5'-D(*CP*CP*CP*AP*AP*GP*CP*GP*GP*C)-3', 5'-D(*GP*CP*CP*GP*CP*TP*TP*GP*GP*G)-3', Poly [ADP-ribose] polymerase 1, ... | | Authors: | Pascal, J.M, Langelier, M.-F. | | Deposit date: | 2010-08-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Poly(ADP-ribose) Polymerase-1 (PARP-1) Zinc Fingers Bound to DNA: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO DNA-DEPENDENT PARP-1 ACTIVITY.
J.Biol.Chem., 286, 2011
|
|
