3CUU
 
 | | Crystal structure of glycogen phosphorylase b in complex with N-(-D-glucopyranosyl)-N'-(2-naphthyl)oxamides | | Descriptor: | Glycogen phosphorylase, muscle form, N-[(naphthalen-1-ylamino)(oxo)acetyl]-beta-D-glucopyranosylamine | | Authors: | Kyritsi, C, Chrysina, E.D, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-04-17 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tracking structure activity relationships of glycogen phosphorylase inhibitors: synthesis, kinetic and crystallographic evaluation of analogues of N-(-D-glucopyranosyl)-N'-oxamides
To be Published
|
|
6FIT
 
 | | FHIT-TRANSITION STATE ANALOG | | Descriptor: | ADENOSINE MONOTUNGSTATE, FRAGILE HISTIDINE TRIAD PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
4MI6
 
 | | Crystal structure of Gpb in complex with SUGAR (N-[4-(5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)BUTANOYL]-BETA-D-GLUCOPYRANOSYLAMINE) | | Descriptor: | Glycogen phosphorylase, muscle form, N-[4-(5,6,7,8-tetrahydronaphthalen-2-yl)butanoyl]-beta-D-glucopyranosylamine | | Authors: | Kantsadi, L.A, Chatzileontiadou, S.M.D, Leonidas, D.D. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines.
Bioorg.Med.Chem., 22, 2014
|
|
1K06
 
 | | Crystallographic Binding Study of 100 mM N-benzoyl-N'-beta-D-glucopyranosyl urea to glycogen phosphorylase b | | Descriptor: | Glycogen Phosphorylase, N-[(phenylcarbonyl)carbamoyl]-beta-D-glucopyranosylamine, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oikonomakos, N.G, Kosmopoulou, M, Zographos, S.E, Leonidas, D.D, Chrysina, E.D, Somsak, L, Nagy, V, Praly, J.P, Docsa, T, Toth, B, Gergely, P. | | Deposit date: | 2001-09-18 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of N-acetyl-N '-beta-D-glucopyranosyl urea and N-benzoyl-N '-beta-D-glucopyranosyl urea to glycogen phosphorylase b: kinetic and crystallographic studies.
Eur.J.Biochem., 269, 2002
|
|
2J9L
 
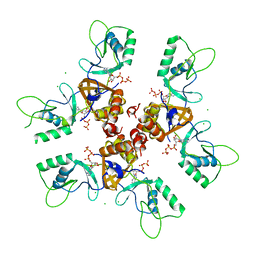 | | Cytoplasmic Domain of the Human Chloride Transporter ClC-5 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE CHANNEL PROTEIN 5, CHLORIDE ION | | Authors: | Meyer, S, Savaresi, S, Forster, I.C, Dutzler, R. | | Deposit date: | 2006-11-13 | | Release date: | 2007-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nucleotide Recognition by the Cytoplasmic Domain of the Human Chloride Transporter Clc-5
Nat.Struct.Mol.Biol., 14, 2006
|
|
1KKF
 
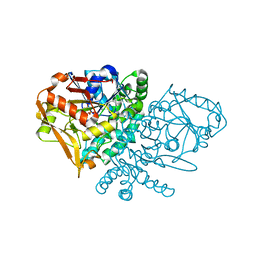 | | Complex of E. coli Adenylosuccinate Synthetase with IMP, Hadacidin, Pyrophosphate, and Mg | | Descriptor: | Adenylosuccinate Synthetase, DIPHOSPHATE, HADACIDIN, ... | | Authors: | Hou, Z, Wang, W, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2001-12-07 | | Release date: | 2002-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | IMP Alone Organizes the Active Site of Adenylosuccinate Synthetase from Escherichia coli.
J.Biol.Chem., 277, 2002
|
|
1L5R
 
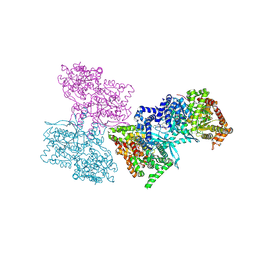 | | Human liver glycogen phosphorylase a complexed with riboflavin, N-Acetyl-beta-D-Glucopyranosylamine and CP-403,700 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, N-acetyl-beta-D-glucopyranosylamine, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Ekstrom, J.L, Pauly, T.A, Carty, M.D, Soeller, W.C, Culp, J, Danley, D.E, Hoover, D.J, Treadway, J.L, Gibbs, E.M, Fletterick, R.J, Day, Y.S.N, Myszka, D.G, Rath, V.L. | | Deposit date: | 2002-03-07 | | Release date: | 2002-12-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity analysis of the purine binding
site of human liver glycogen phosphorylase.
Chem.Biol., 9, 2002
|
|
3CZQ
 
 | | Crystal structure of putative polyphosphate kinase 2 from Sinorhizobium meliloti | | Descriptor: | FORMIC ACID, GLYCEROL, Putative polyphosphate kinase 2 | | Authors: | Osipiuk, J, Evdokimova, E, Nocek, B, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Polyphosphate-dependent synthesis of ATP and ADP by the family-2 polyphosphate kinases in bacteria.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3CRZ
 
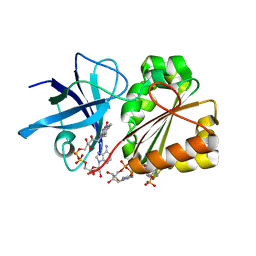 | | Ferredoxin-NADP Reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP+ reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Han, H, Schonbrunn, E. | | Deposit date: | 2008-04-08 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystallographic and solution state nuclear magnetic resonance spectroscopic investigations of NADP+ binding to ferredoxin NADP reductase from Pseudomonas aeruginosa
Biochemistry, 47, 2008
|
|
3CUT
 
 | | Crystal structure of glycogen phosphorylase b in complex with N-(-D-glucopyranosyl)-N'-(2-naphthyl)oxamide | | Descriptor: | Glycogen phosphorylase, muscle form, N-[(naphthalen-2-ylamino)(oxo)acetyl]-beta-D-glucopyranosylamine | | Authors: | Kyritsi, C, Chrysina, E.D, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-04-17 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tracking structure activity relationships of glycogen phosphorylase inhibitors: synthesis, kinetic and crystallographic evaluation of analogues of N-(-D-glucopyranosyl)-N'-oxamides
To be Published
|
|
3FJY
 
 | |
2GJ4
 
 | | Structure of rabbit muscle glycogen phosphorylase in complex with ligand | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 2-CHLORO-N-[(1R,2R)-1-HYDROXY-2,3-DIHYDRO-1H-INDEN-2-YL]-6H-THIENO[2,3-B]PYRROLE-5-CARBOXAMIDE, Glycogen phosphorylase, ... | | Authors: | Otterbein, L.R, Pannifer, A.D, Tucker, J, Breed, J, Oikonomakos, N.G, Rowsell, S, Pauptit, R.A, Claire, M. | | Deposit date: | 2006-03-30 | | Release date: | 2007-02-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel thienopyrrole glycogen phosphorylase inhibitors: synthesis, in vitro SAR and crystallographic studies.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4MB6
 
 | | Crystal structure of adenine phosphoribosyltransferase from Yersinia pseudotuberculosis. | | Descriptor: | Adenine phosphoribosyltransferase, SODIUM ION | | Authors: | Pavithra, G.C, Kim, J, Hegde, R.P, Almo, S.C, Ramagopal, U.A, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of adenine phosphoribosyltransferase from Yersinia pseudotuberculosis
To be published
|
|
3CUW
 
 | | Crystal structure of glycogen phosphorylase b in complex with N-(-D-glucopyranosyl)-N'-(2-naphthyl)oxamides | | Descriptor: | Glycogen phosphorylase, muscle form, N-[oxo(phenylamino)acetyl]-beta-D-glucopyranosylamine | | Authors: | Kyritsi, C, Chrysina, E.D, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-04-17 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tracking structure activity relationships of glycogen phosphorylase inhibitors: synthesis, kinetic and crystallographic evaluation of analogues of N-(-D-glucopyranosyl)-N'-oxamides
To be Published
|
|
1KWA
 
 | | HUMAN CASK/LIN-2 PDZ DOMAIN | | Descriptor: | HCASK/LIN-2 PROTEIN, SULFATE ION | | Authors: | Daniels, D.L, Cohen, A.R, Anderson, J.M, Brunger, A.T. | | Deposit date: | 1998-01-16 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of the hCASK PDZ domain reveals the structural basis of class II PDZ domain target recognition
Nat.Struct.Biol., 5, 1998
|
|
1KRM
 
 | |
1L5S
 
 | | Human liver glycogen phosphorylase complexed with uric acid, N-Acetyl-beta-D-glucopyranosylamine, and CP-403,700 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Glycogen phosphorylase, liver form, ... | | Authors: | Ekstrom, J.L, Pauly, T.A, Carty, M.D, Soeller, W.C, Culp, J, Danley, D.E, Hoover, D.J, Treadway, J.L, Gibbs, E.M, Fletterick, R.J, Day, Y.S.N, Myszka, D.G, Rath, V.L. | | Deposit date: | 2002-03-07 | | Release date: | 2002-12-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity analysis of the purine binding
site of human liver glycogen phosphorylase.
Chem.Biol., 9, 2002
|
|
1L5Q
 
 | | Human liver glycogen phosphorylase a complexed with caffeine, N-Acetyl-beta-D-glucopyranosylamine, and CP-403700 | | Descriptor: | CAFFEINE, Glycogen phosphorylase, liver form, ... | | Authors: | Ekstrom, J.L, Pauly, T.A, Carty, M.D, Soeller, W.C, Culp, J, Danley, D.E, Hoover, D.J, Treadway, J.L, Gibbs, E.M, Fletterick, R.J, Day, Y.S.N, Myszka, D.G, Rath, V.L. | | Deposit date: | 2002-03-07 | | Release date: | 2002-12-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-activity analysis of the purine binding
site of human liver glycogen phosphorylase.
Chem.Biol., 9, 2002
|
|
4KHA
 
 | | Structural basis of histone H2A-H2B recognition by the essential chaperone FACT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Histone H2A, ... | | Authors: | Hondele, M, Halbach, F, Hassler, M, Ladurner, A.G. | | Deposit date: | 2013-04-30 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of histone H2A-H2B recognition by the essential chaperone FACT.
Nature, 499, 2013
|
|
1KKB
 
 | | Complex of Escherichia coli Adenylosuccinate Synthetase with IMP and Hadacidin | | Descriptor: | Adenylosuccinate Synthetase, HADACIDIN, INOSINIC ACID | | Authors: | Hou, Z, Wang, W, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2001-12-06 | | Release date: | 2002-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | IMP Alone Organizes the Active Site of Adenylosuccinate Synthetase from Escherichia coli.
J.Biol.Chem., 277, 2002
|
|
1KTI
 
 | | BINDING OF 100 MM N-ACETYL-N'-BETA-D-GLUCOPYRANOSYL UREA TO GLYCOGEN PHOSPHORYLASE B: KINETIC AND CRYSTALLOGRAPHIC STUDIES | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, N-(acetylcarbamoyl)-beta-D-glucopyranosylamine, ... | | Authors: | Oikonomakos, N.G, Kosmopoulou, M, Zographos, S.E, Leonidas, D.D, Chrysina, E.D, Somsak, L, Nagy, V, Praly, J.P, Docsa, T, Toth, B, Gergely, P. | | Deposit date: | 2002-01-16 | | Release date: | 2002-01-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Binding of N-acetyl-N'-beta-D-glucopyranosyl urea and N-benzoyl-N'-beta-D-glucopyranosyl urea to glycogen phosphorylase b: kinetic and crystallographic studies.
Eur.J.Biochem., 269, 2002
|
|
2JA3
 
 | | Cytoplasmic Domain of the Human Chloride Transporter ClC-5 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE CHANNEL PROTEIN 5 | | Authors: | Meyer, S, Savaresi, S, Forster, I.C, Dutzler, R. | | Deposit date: | 2006-11-21 | | Release date: | 2007-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Nucleotide Recognition by the Cytoplasmic Domain of the Human Chloride Transporter Clc-5
Nat.Struct.Mol.Biol., 14, 2006
|
|
6E0N
 
 | | Structure of Elizabethkingia meningoseptica CdnE cyclic dinucleotide synthase with GTP and Apcpp | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
4MI3
 
 | | Crystal structure of Gpb in complex with SUGAR (N-{(2R)-2-METHYL-3-[4-(PROPAN-2-YL)PHENYL]PROPANOYL}-BETA-D-GLUCOPYRANOSYLAMINE) (S21) | | Descriptor: | Glycogen phosphorylase, muscle form, N-{(2R)-2-methyl-3-[4-(propan-2-yl)phenyl]propanoyl}-beta-D-glucopyranosylamine | | Authors: | Kantsadi, L.A, Chatzileontiadou, S.M.D, Leonidas, D.D. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines.
Bioorg.Med.Chem., 22, 2014
|
|
6GYF
 
 | | Crystal structure of NadR protein in complex with NR | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MAGNESIUM ION, Nicotinamide-nucleotide adenylyltransferase NadR family / Ribosylnicotinamide kinase, ... | | Authors: | Singh, R, Stetsenko, A, Jaehme, M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2018-06-29 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Characterization of NadR fromLactococcus lactis.
Molecules, 25, 2020
|
|
