6TYN
 
 | | Salmonella Typhi PltB Homopentamer with Neu-5NAc-9OAc-alpha-2-3-Gal-beta-1-4-GlcNAc Glycans | | Descriptor: | 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pertussis like toxin subunit B | | Authors: | Nguyen, T, Milano, S.K, Yang, Y.A, Song, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The role of 9-O-acetylated glycan receptor moieties in the typhoid toxin binding and intoxication.
Plos Pathog., 16, 2020
|
|
5DPX
 
 | | 1,2,4-Triazole-3-thione compounds as inhibitors of L1, di-zinc metallo-beta-lactamases. | | Descriptor: | 5-(2-methylphenyl)-3H-1,2,4-triazole-3-thione, Metallo-beta-lactamase L1 type 3, SULFATE ION, ... | | Authors: | Nauton, L, Garau, G, Khan, R, Dideberg, O. | | Deposit date: | 2015-09-14 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1,2,4-Triazole-3-thione Compounds as Inhibitors of Dizinc Metallo-beta-lactamases.
ChemMedChem, 12, 2017
|
|
7UYO
 
 | |
7UYN
 
 | |
7UYP
 
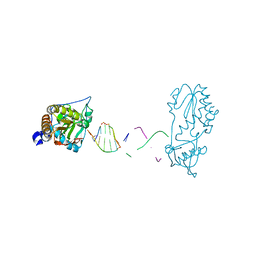 | |
5FWT
 
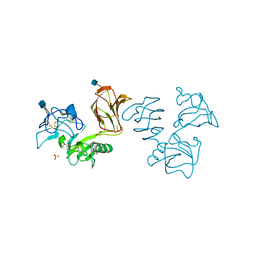 | | Wnt modulator Kremen crystal form I at 2.10A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, KREMEN PROTEIN 1, ... | | Authors: | Zebisch, M, Jackson, V.A, Jones, E.Y. | | Deposit date: | 2016-02-21 | | Release date: | 2016-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Structure, 24, 2016
|
|
5DX8
 
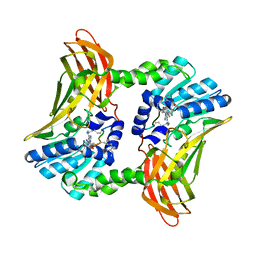 | |
6I99
 
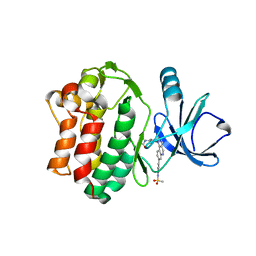 | | Bone Marrow Tyrosine Kinase in Chromosome X in complex with a newly designed covalent inhibitor JS24 | | Descriptor: | Cytoplasmic tyrosine-protein kinase BMX, ~{N}-[2-methyl-5-[8-[4-(methylsulfonylamino)phenyl]-2-oxidanylidene-benzo[h][1,6]naphthyridin-1-yl]phenyl]-3-oxidanyl-propanamide | | Authors: | Sousa, B.B, Matias, P.M, Marques, M.C, Seixas, J.D, Bernardes, G.J.L. | | Deposit date: | 2018-11-22 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural and biophysical insights into the mode of covalent binding of rationally designed potent BMX inhibitors
Chem.Biol., 2020
|
|
8I8N
 
 | | Crystal structure of the ternary complex of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with Dephosphocoenzyme-A and Phosphonoacetic acid at 2.22 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, DEPHOSPHO COENZYME A, GLYCEROL, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2023-02-04 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of the ternary complex of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with Dephosphocoenzyme-A and Phosphonoacetic acid at 2.22 A resolution.
To Be Published
|
|
6E8W
 
 | | MPER-TM Domain of HIV-1 envelope glycoprotein (Env) | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Fu, Q, Shaik, M.M, Cai, Y, Ghantous, F, Piai, A, Peng, H, Rits-Volloch, S, Liu, Z, Harrison, S.C, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2018-07-31 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane proximal external region of HIV-1 envelope glycoprotein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1F2L
 
 | |
6IVY
 
 | | Crystal structure of iron-bound HitA from Pseudomonas aeruginosa | | Descriptor: | FE (III) ION, PHOSPHATE ION, Periplasmic Ferric iron-binding Protein HitA | | Authors: | Zhang, Z.R, Li, H.Y. | | Deposit date: | 2018-12-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9999994 Å) | | Cite: | Identification and Characterization of a Metalloprotein Involved in Gallium Internalization in Pseudomonas aeruginosa.
Acs Infect Dis., 5, 2019
|
|
1BD0
 
 | | ALANINE RACEMASE COMPLEXED WITH ALANINE PHOSPHONATE | | Descriptor: | ALANINE RACEMASE, {1-[(3-HYDROXY-METHYL-5-PHOSPHONOOXY-METHYL-PYRIDIN-4-YLMETHYL)-AMINO]-ETHYL}-PHOSPHONIC ACID | | Authors: | Stamper, G.F, Morollo, A.A, Ringe, D. | | Deposit date: | 1998-05-12 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Reaction of alanine racemase with 1-aminoethylphosphonic acid forms a stable external aldimine.
Biochemistry, 37, 1998
|
|
4L1X
 
 | | Crystal Structuer of Human 3-alpha Hydroxysteroid Dehydrogenase Type 3 V54L Mutant in Complex with NADP+ and Progesterone | | Descriptor: | Aldo-keto reductase family 1 member C2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROGESTERONE, ... | | Authors: | Zhang, B, Hu, X.-J, Lin, S.-X. | | Deposit date: | 2013-06-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human 3-alpha hydroxysteroid dehydrogenase type 3 (3 alpha-HSD3): The V54L mutation restricting the steroid alternative binding and enhancing the 20 alpha-HSD activity
J.Steroid Biochem.Mol.Biol., 141, 2014
|
|
4XHM
 
 | |
6OZ4
 
 | |
8RTH
 
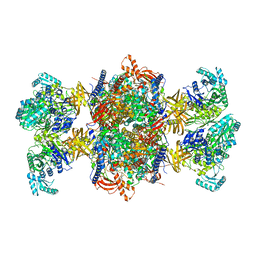 | | Trypanosoma brucei 3-methylcrotonyl-CoA carboxylase | | Descriptor: | 3-methylcrotonyl-CoA carboxylase, putative, 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ... | | Authors: | Ruiz, F.M, Plaza-Pegueroles, A, Fernandez-Tornero, C. | | Deposit date: | 2024-01-26 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | The cryo-EM structure of trypanosome 3-methylcrotonyl-CoA carboxylase provides mechanistic and dynamic insights into its enzymatic function.
Structure, 2024
|
|
6OZ2
 
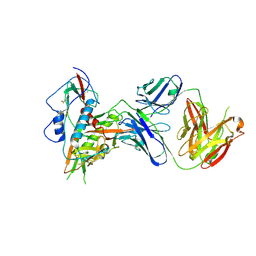 | |
4XXW
 
 | |
6PH2
 
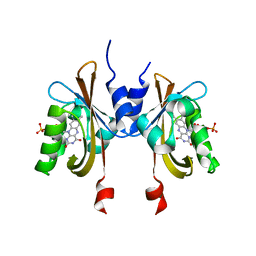 | | Complete LOV domain from the LOV-HK sensory protein from Brucella abortus (mutant C69S, construct 15-155) | | Descriptor: | Blue-light-activated histidine kinase, FLAVIN MONONUCLEOTIDE | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
5OH4
 
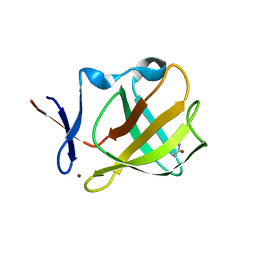 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Piperidine-2,6-dione (Glutarimide) | | Descriptor: | Cereblon isoform 4, ZINC ION, piperidine-2,6-dione | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
6N1K
 
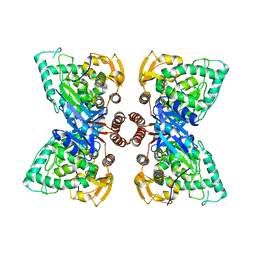 | |
5OH2
 
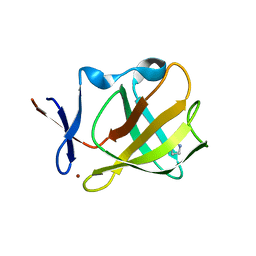 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Pyrrolidin-2-one (Butyrolactam) | | Descriptor: | Cereblon isoform 4, ZINC ION, pyrrolidin-2-one | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
5OHB
 
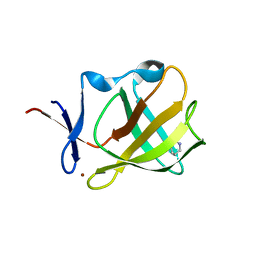 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Piperidin-2-one (Valerolactam) | | Descriptor: | Cereblon isoform 4, ZINC ION, piperidin-2-one | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
1FNS
 
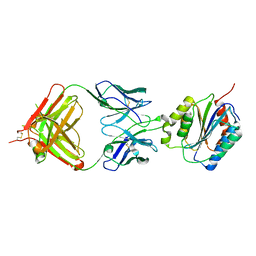 | |
