3L6X
 
 | | Crystal structure of p120 catenin in complex with E-cadherin | | Descriptor: | Catenin delta-1, E-cadherin, SULFATE ION | | Authors: | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | Deposit date: | 2009-12-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
3HSF
 
 | | HEAT SHOCK TRANSCRIPTION FACTOR (HSF) | | Descriptor: | HEAT SHOCK TRANSCRIPTION FACTOR | | Authors: | Damberger, F.F, Pelton, J.G, Liu, C, Cho, H, Harrison, C.J, Nelson, H.C.M, Wemmer, D.E. | | Deposit date: | 1995-08-07 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure and dynamics of the DNA-binding domain of the heat shock factor from Kluyveromyces lactis.
J.Mol.Biol., 254, 1995
|
|
3HMS
 
 | |
3E1U
 
 | | The Crystal Structure of the Anti-Viral APOBEC3G Catalytic Domain | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Holden, L, Prochnow, C, Chang, Y.P, Bransteitter, R, Chelico, L, Sen, U, Stevens, R.C, Goodman, R.F, Chen, X.S. | | Deposit date: | 2008-08-04 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications.
Nature, 456, 2008
|
|
3E4Z
 
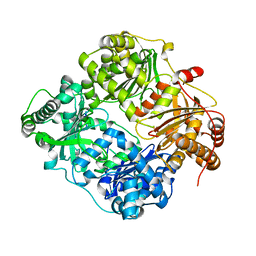 | | Crystal structure of human insulin degrading enzyme in complex with insulin-like growth factor II | | Descriptor: | Insulin-degrading enzyme, Insulin-like growth factor II, ZINC ION | | Authors: | Guo, Q, Manolopoulou, M, Tang, W.-J. | | Deposit date: | 2008-08-12 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Molecular Basis for the Recognition and Cleavages of IGF-II, TGF-alpha, and Amylin by Human Insulin-Degrading Enzyme.
J.Mol.Biol., 395, 2010
|
|
3EBE
 
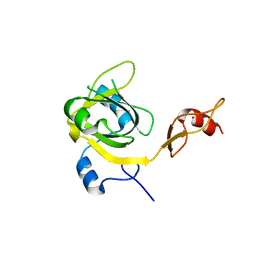 | |
3HMT
 
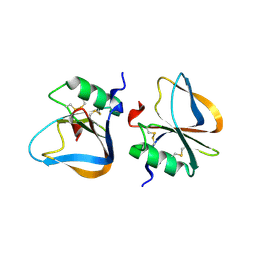 | |
3LH0
 
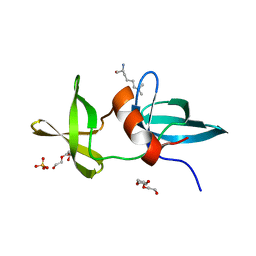 | |
3LLO
 
 | | Crystal structure of the STAS domain of motor protein prestin (anion transporter SLC26A5) | | Descriptor: | Prestin, octyl beta-D-glucopyranoside | | Authors: | Pasqualetto, E, Aiello, R, Bonetto, G, Battistutta, R. | | Deposit date: | 2010-01-29 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure of the cytosolic portion of the motor protein prestin and functional role of the STAS domain in SLC26/SulP anion transporters.
J.Mol.Biol., 400, 2010
|
|
3LHC
 
 | | Crystal structure of cyanovirin-n swapping domain b mutant | | Descriptor: | Cyanovirin-N, PHOSPHATE ION, SODIUM ION | | Authors: | Matei, E, Zheng, A, Furey, W, Rose, J, Aiken, C, Gronenborn, A.M. | | Deposit date: | 2010-01-21 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Anti-HIV activity of defective cyanovirin-N mutants is restored by dimerization.
J.Biol.Chem., 285, 2010
|
|
3KKO
 
 | | Crystal structure of M-Ras P40D/D41E/L51R in complex with GppNHp | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein M-Ras, ... | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
3DWH
 
 | |
3E07
 
 | | Crystal structure of spatzle cystine knot | | Descriptor: | GLYCEROL, Protein spaetzle | | Authors: | Hoffmann, A, Funkner, A, Neumann, P, Juhnke, S, Walther, M, Schierhorn, A, Weininger, U, Balbach, J, Reuter, G, Stubbs, M.T. | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biophysical Characterization of Refolded Drosophila Spatzle, a Cystine Knot Protein, Reveals Distinct Properties of Three Isoforms
J.Biol.Chem., 283, 2008
|
|
3KO0
 
 | | Structure of the tfp-ca2+-bound activated form of the s100a4 Metastasis factor | | Descriptor: | 10-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPYL]-2-TRIFLUOROMETHYL-10H-PHENOTHIAZINE, CALCIUM ION, Protein S100-A4 | | Authors: | Malashkevich, V.N, Dulyaninova, N.G, Knight, D, Almo, S.C, Bresnick, A.R. | | Deposit date: | 2009-11-12 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phenothiazines inhibit S100A4 function by inducing protein oligomerization.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KVR
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | 2,5-anhydro-4-deoxy-D-erythro-pent-4-enitol, 5-FLUOROURACIL, SULFATE ION, ... | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
3GMH
 
 | | Crystal Structure of the Mad2 Dimer | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2A, SULFATE ION | | Authors: | Ozkan, E, Luo, X, Machius, M, Yu, H, Deisenhofer, J. | | Deposit date: | 2009-03-13 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of an intermediate conformer of the spindle checkpoint protein Mad2.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3KZD
 
 | |
3HCM
 
 | | Crystal structure of human S100B in complex with S45 | | Descriptor: | (3R)-3-[3-(4-chlorophenyl)-1,2,4-oxadiazol-5-yl]piperidine, ACETATE ION, CALCIUM ION, ... | | Authors: | Mangani, S, Cesari, L. | | Deposit date: | 2009-05-06 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragmenting the S100B-p53 Interaction: Combined Virtual/Biophysical Screening Approaches to Identify Ligands
Chemmedchem, 5, 2010
|
|
3EVS
 
 | | Crystal structure of the GDF-5:BMP receptor IB complex. | | Descriptor: | Bone morphogenetic protein receptor type-1B, Growth/differentiation factor 5 | | Authors: | Kotzsch, A, Mueller, T.D. | | Deposit date: | 2008-10-13 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure analysis reveals a spring-loaded latch as molecular mechanism for GDF-5-type I receptor specificity.
Embo J., 28, 2009
|
|
3LI6
 
 | | Crystal structure and trimer-monomer transition of N-terminal domain of EhCaBP1 from Entamoeba histolytica | | Descriptor: | CALCIUM ION, Calcium-binding protein | | Authors: | Kumar, S, Ahmad, E, Kumar, S, Mansuri, M.S, Khan, R.H, Samudrala, G. | | Deposit date: | 2010-01-24 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structure and trimer-monomer transition of N-terminal domain of EhCaBP1 from Entamoeba histolytica
Biophys.J., 98, 2010
|
|
3LJW
 
 | | Crystal Structure of the Second Bromodomain of Human Polybromo | | Descriptor: | ACETATE ION, Protein polybromo-1, SODIUM ION | | Authors: | Charlop-Powers, Z, Zhou, M.M, Zeng, L, Zhang, Q. | | Deposit date: | 2010-01-26 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structural insights into selective histone H3 recognition by the human Polybromo bromodomain 2.
Cell Res., 20, 2010
|
|
3F0L
 
 | | Crystal structure of oxidized D105N Synechocystis sp. PcyA | | Descriptor: | 1,2-ETHANEDIOL, BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Kohler, A.C, Gae, D.D. | | Deposit date: | 2008-10-25 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for hydration dynamics in radical stabilization of bilin reductase mutants.
Biochemistry, 49, 2010
|
|
3LB9
 
 | | Crystal structure of the B. circulans cpA123 circular permutant | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | D'Angelo, I, Reitinger, S, Ludwiczek, M, Strynadka, N, Withers, S.G, Mcintosh, L.P. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Circular permutation of Bacillus circulans xylanase: a kinetic and structural study.
Biochemistry, 49, 2010
|
|
3F2O
 
 | | Crystal Structure of human splA/ryanodine receptor domain and SOCS box containing 1 (SPSB1) in complex with a 20-residue VASA peptide | | Descriptor: | 20-mer peptide from ATP-dependent RNA helicase vasa, SPRY domain-containing SOCS box protein 1 | | Authors: | Filippakopoulos, P, Sharpe, T, Keates, T, Murray, J.W, Savitsky, P, Roos, A, Pike, A.C.W, Von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for Par-4 recognition by the SPRY domain- and SOCS box-containing proteins SPSB1, SPSB2, and SPSB4.
J.Mol.Biol., 401, 2010
|
|
3KU4
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | SULFATE ION, Uridine phosphorylase | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-26 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
