5YBG
 
 | | Crystal structure of the GluA2o LBD in complex with glutamate and LY451395 | | Descriptor: | ACETATE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2017-09-04 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | HBT1, a Novel AMPA Receptor Potentiator with Lower Agonistic Effect, Avoided Bell-Shaped Response in In Vitro BDNF Production.
J. Pharmacol. Exp. Ther., 364, 2018
|
|
4U6Q
 
 | | CtBP1 bound to inhibitor 2-(hydroxyimino)-3-phenylpropanoic acid | | Descriptor: | (2E)-2-(hydroxyimino)-3-phenylpropanoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C-terminal-binding protein 1, ... | | Authors: | Hilbert, B.J, Morris, B.L, Ellis, K.C, Paulsen, J.L, Schiffer, C.A, Grossman, S.R, Royer Jr, W.E. | | Deposit date: | 2014-07-29 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Guided Design of a High Affinity Inhibitor to Human CtBP.
Acs Chem.Biol., 10, 2015
|
|
5YDG
 
 | | Crystal structure of the Arabidopsis thaliana chloroplast RNA editing factors 2(MORF2) | | Descriptor: | Multiple organellar RNA editing factor 2, chloroplastic | | Authors: | Wang, X, Yang, J.Y, Wang, Y.L, Gao, Y.S. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Crystal structure of the chloroplast RNA editing factor MORF2
Biochem. Biophys. Res. Commun., 495, 2018
|
|
4U6X
 
 | | Crystal Structure of HLA-A*0201 in complex with ALQDA, a 15 mer self-peptide | | Descriptor: | ALQDA peptide, ALQDAGDSSRKEYFI, Beta-2-microglobulin, ... | | Authors: | Gras, S, Chabrol, E, Rossjohn, J. | | Deposit date: | 2014-07-30 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Naturally Processed Non-canonical HLA-A*02:01 Presented Peptides.
J.Biol.Chem., 290, 2015
|
|
4U73
 
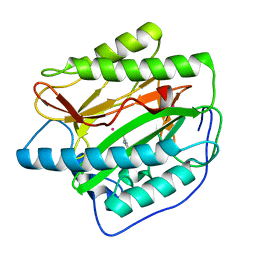 | |
5YDM
 
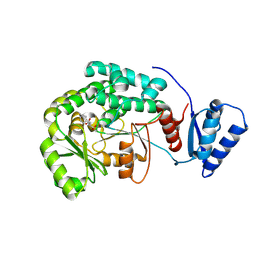 | |
5YDV
 
 | | Regulatory domain of HypT from Salmonella typhimurium complexed with HOCl (HOCl-bound form) | | Descriptor: | Cell density-dependent motility repressor, SULFATE ION, hypochlorous acid | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-14 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4U79
 
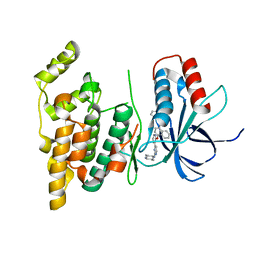 | | Crystal structure of human JNK3 in complex with a benzenesulfonamide inhibitor. | | Descriptor: | Mitogen-activated protein kinase 10, N-{4-[(3-{2-[(trans-4-aminocyclohexyl)amino]pyrimidin-4-yl}pyridin-2-yl)oxy]naphthalen-1-yl}benzenesulfonamide | | Authors: | Mohr, C. | | Deposit date: | 2014-07-30 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Unfolded Protein Response in Cancer: IRE1 alpha Inhibition by Selective Kinase Ligands Does Not Impair Tumor Cell Viability.
Acs Med.Chem.Lett., 6, 2015
|
|
5YCB
 
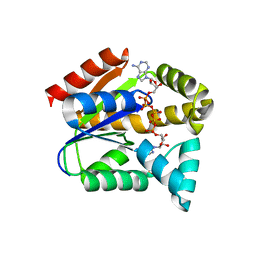 | |
4U7I
 
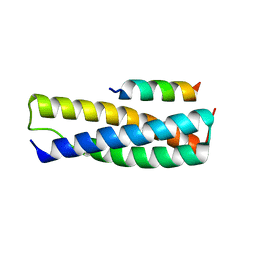 | | Structure of the complex of Spartin MIT and IST1 MIM | | Descriptor: | IST1 homolog, Spartin | | Authors: | Guo, E.Z, Xu, Z. | | Deposit date: | 2014-07-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Distinct Mechanisms of Recognizing Endosomal Sorting Complex Required for Transport III (ESCRT-III) Protein IST1 by Different Microtubule Interacting and Trafficking (MIT) Domains.
J.Biol.Chem., 290, 2015
|
|
5YCP
 
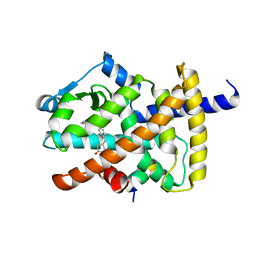 | | Human PPARgamma ligand binding domain complexed with Rosiglitazone | | Descriptor: | 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), Nuclear receptor coactivator 1, ... | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2017-09-07 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Enhanced Anti-Diabetic Efficacy of Lobeglitazone on PPAR gamma.
Sci Rep, 8, 2018
|
|
4U7K
 
 | | Crystal structure of the Clostridium histolyticum colH collagenase polycystic kidney disease-like domain 2a in the presence of calcium at 1.9 Angstrom resolution | | Descriptor: | CALCIUM ION, ColH protein | | Authors: | Bauer, R, Janowska, K, Sakon, J, Matsushita, O, Latimer, E. | | Deposit date: | 2014-07-31 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structures of three polycystic kidney disease-like domains from Clostridium histolyticum collagenases ColG and ColH.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5YD0
 
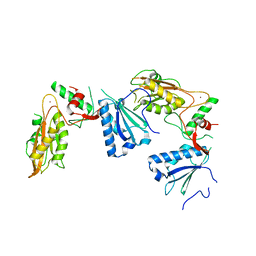 | | Crystal structure of Schlafen 13 (SLFN13) N'-domain | | Descriptor: | Schlafen 8, ZINC ION | | Authors: | Yang, J.-Y, Gao, S. | | Deposit date: | 2017-09-09 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.182 Å) | | Cite: | Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control
Nat Commun, 9, 2018
|
|
4U84
 
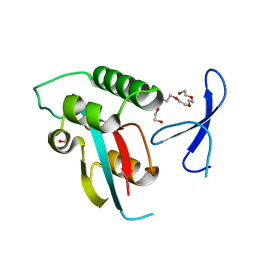 | | Human Pin1 with S-hydroxyl-cysteine 113 | | Descriptor: | POLYETHYLENE GLYCOL (N=34), Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Li, W, Zhang, Y. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer's disease.
Neurobiol.Dis., 76, 2015
|
|
5YE9
 
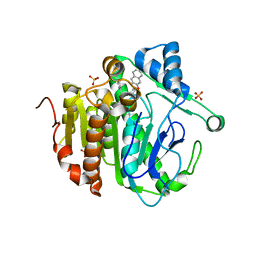 | | The crystal structure of Lp-PLA2 in complex with a novel inhibitor | | Descriptor: | N-[4-[(3-cyano-4-naphthalen-2-yloxy-phenyl)sulfamoyl]phenyl]ethanamide, Platelet-activating factor acetylhydrolase, SULFATE ION | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2017-09-15 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | Structure-Guided Discovery of Novel, Potent, and Orally Bioavailable Inhibitors of Lipoprotein-Associated Phospholipase A2.
J. Med. Chem., 60, 2017
|
|
4U89
 
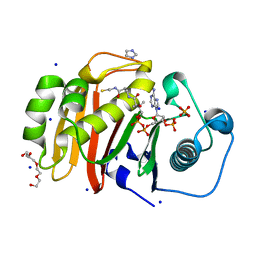 | |
5YEQ
 
 | | The structure of Sac-KARI protein | | Descriptor: | 1,2-ETHANEDIOL, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, ... | | Authors: | Ko, T.P, Chen, C.Y, Lin, K.F, Lin, B.L, Huang, C.H, Chiang, C.H, Horng, J.C, Tsai, M.D. | | Deposit date: | 2017-09-19 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | NADH/NADPH bi-cofactor-utilizing and thermoactive ketol-acid reductoisomerase from Sulfolobus acidocaldarius
Sci Rep, 8, 2018
|
|
4U8J
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Y104A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U91
 
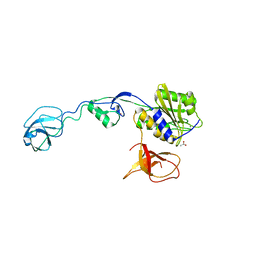 | | GephE in complex with Para-Phenyl crosslinked Glycine receptor beta subunit derived dimeric peptide | | Descriptor: | 1,1'-benzene-1,4-diylbis(1H-pyrrole-2,5-dione), 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Kasaragod, V.B, Maric, H.M, Schindelin, H. | | Deposit date: | 2014-08-05 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of High-Affinity Dimeric Inhibitors Targeting the Interactions between Gephyrin and Inhibitory Neurotransmitter Receptors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5YEZ
 
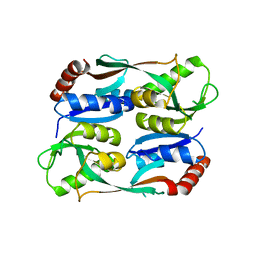 | | Regulatory domain of HypT M206Q mutant from Salmonella typhimurium | | Descriptor: | Cell density-dependent motility repressor | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-20 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4U9B
 
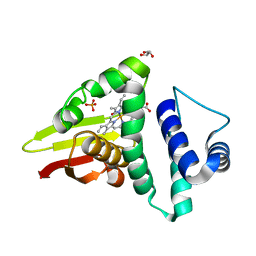 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II)NO ligation state | | Descriptor: | GLYCEROL, NITRIC OXIDE, NO-binding heme-dependent sensor protein, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5YDU
 
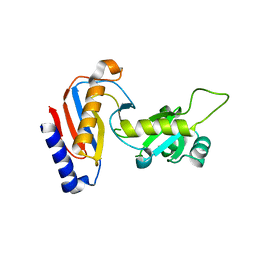 | | Crystal structure of Utp30 | | Descriptor: | PHOSPHATE ION, Ribosome biogenesis protein UTP30 | | Authors: | Hu, J, Zhu, X, Ye, K. | | Deposit date: | 2017-09-14 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.646 Å) | | Cite: | Structure and RNA recognition of ribosome assembly factor Utp30.
RNA, 23, 2017
|
|
5YE7
 
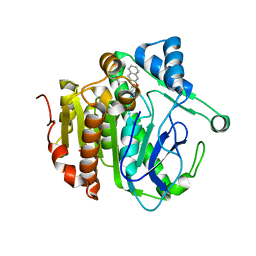 | | The crystal structure of Lp-PLA2 in complex with a novel inhibitor | | Descriptor: | N-[4-[(4-naphthalen-2-yloxyphenyl)sulfamoyl]phenyl]ethanamide, Platelet-activating factor acetylhydrolase, SULFATE ION | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2017-09-15 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Structure-Guided Discovery of Novel, Potent, and Orally Bioavailable Inhibitors of Lipoprotein-Associated Phospholipase A2.
J. Med. Chem., 60, 2017
|
|
4U9H
 
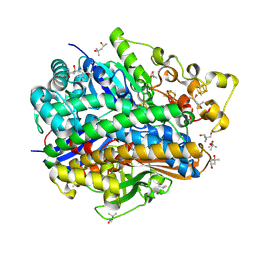 | | Ultra High Resolution Structure Of The Ni-R State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Nishikawa, K, Lubitz, W. | | Deposit date: | 2014-08-06 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Hydrogens detected by subatomic resolution protein crystallography in a [NiFe] hydrogenase.
Nature, 520, 2015
|
|
5YEJ
 
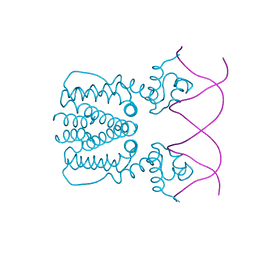 | | Crystal structure of BioQ with its naturel double-stranded DNA operator | | Descriptor: | DNA (5'-D(*AP*CP*CP*TP*GP*AP*AP*CP*AP*CP*CP*GP*TP*TP*CP*AP*AP*GP*T)-3'), DNA (5'-D(*AP*CP*TP*TP*GP*AP*AP*CP*GP*GP*TP*GP*TP*TP*CP*AP*GP*GP*T)-3'), TetR family transcriptional regulator | | Authors: | Yan, L, Guan, Z.Y, Zou, T.T. | | Deposit date: | 2017-09-17 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structural insights into operator recognition by BioQ in the Mycobacterium smegmatis biotin synthesis pathway.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
