3MF0
 
 | | Crystal structure of PDE5A GAF domain (89-518) | | Descriptor: | cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Wang, H, Robinson, H, Ke, H. | | Deposit date: | 2010-04-01 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain.
J.Biol.Chem., 285, 2010
|
|
3MMN
 
 | | Crystal structure of the receiver domain of the histidine kinase CKI1 from Arabidopsis thaliana complexed with Mg2+ | | Descriptor: | Histidine kinase homolog, MAGNESIUM ION | | Authors: | Marek, J, Klumpler, T, Pekarova, B, Triskova, O, Horak, J, Zidek, L, Dopitova, R, Hejatko, J, Janda, L. | | Deposit date: | 2010-04-20 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and binding specificity of the receiver domain of sensor histidine kinase CKI1 from Arabidopsis thaliana.
Plant J., 67, 2011
|
|
3M5C
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase K302E mutant complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Reversible post-translational carboxylation modulates the enzymatic activity of N-acetyl-L-ornithine transcarbamylase.
Biochemistry, 49, 2010
|
|
3MPN
 
 | | F177R1 mutant of LeuT | | Descriptor: | CHLORIDE ION, LEUCINE, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, ... | | Authors: | Kroncke, B.M. | | Deposit date: | 2010-04-27 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural origins of nitroxide side chain dynamics on membrane protein alpha-helical sites.
Biochemistry, 49, 2010
|
|
3MWV
 
 | | Crystal structure of HCV NS5B polymerase | | Descriptor: | Genome polyprotein | | Authors: | Coulombe, R. | | Deposit date: | 2010-05-06 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Importance of ligand bioactive conformation in the discovery of potent indole-diamide inhibitors of the hepatitis C virus NS5B.
J.Am.Chem.Soc., 132, 2010
|
|
3MWW
 
 | | Crystal structure of HCV NS5B polymerase | | Descriptor: | 1-[2-(4-carboxypiperidin-1-yl)-2-oxoethyl]-3-cyclohexyl-2-furan-3-yl-1H-indole-6-carboxylic acid, Genome polyprotein, SULFATE ION | | Authors: | Coulombe, R. | | Deposit date: | 2010-05-06 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Importance of ligand bioactive conformation in the discovery of potent indole-diamide inhibitors of the hepatitis C virus NS5B.
J.Am.Chem.Soc., 132, 2010
|
|
3PA0
 
 | | Crystal Structure of Chiral Gamma-PNA with Complementary DNA Strand: Insight Into the Stability and Specificity of Recognition an Conformational Preorganization | | Descriptor: | 1,2-ETHANEDIOL, DNA 5'-D(*AP*TP*CP*TP*GP*TP*GP*GP*TP*C)-3', PEPTIDE NUCLEIC ACID, ... | | Authors: | Yeh, J.I, Shivachev, B, Du, S. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of chiral gammaPNA with complementary DNA strand: insights into the stability and specificity of recognition and conformational preorganization.
J.Am.Chem.Soc., 132, 2010
|
|
3OWS
 
 | |
3OX7
 
 | | The crystal structure of uPA complex with peptide inhibitor MH027 at pH4.6 | | Descriptor: | MH027, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Jiang, L.G, Andreasen, P.A, Huang, M.D. | | Deposit date: | 2010-09-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The binding mechanism of a peptidic cyclic serine protease inhibitor
J.Mol.Biol., 412, 2011
|
|
3P4K
 
 | | The third conformation of p38a MAP kinase observed in phosphorylated p38a and in solution | | Descriptor: | MAP kinase 14, Mitogen-activated protein kinase 14 | | Authors: | Akella, R, Min, X, Wu, Q, Gardner, K.H, Goldsmith, E.J. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | The third conformation of p38a MAP kinase observed in phosphorylated p38a and in solution
Structure, 18, 2010
|
|
4H5V
 
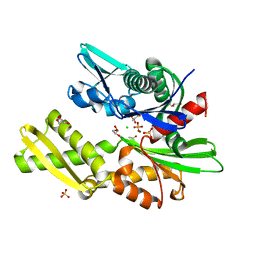 | | HSC70 NBD with Mg | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stec, B. | | Deposit date: | 2012-09-18 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | New crystal structures of HSC-70 ATP binding domain confirm the role of individual binding pockets and suggest a new method of inhibition.
Biochimie, 108, 2015
|
|
3P7J
 
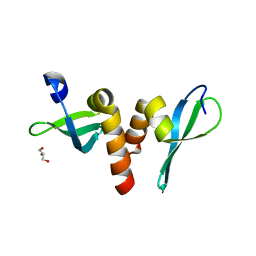 | | Drosophila HP1a chromo shadow domain | | Descriptor: | GLYCEROL, Heterochromatin protein 1 | | Authors: | Kim, D, Chruszcz, M, Minor, W, Khorasanizadeh, S. | | Deposit date: | 2010-10-12 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The HP1a Disordered C Terminus and Chromo Shadow Domain Cooperate to Select Target Peptide Partners.
Chembiochem, 12, 2011
|
|
3N1W
 
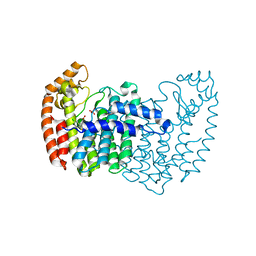 | | Human FPPS COMPLEX WITH FBS_02 | | Descriptor: | (5-chloro-1-benzothiophen-3-yl)acetic acid, FARNESYL PYROPHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Rondeau, J.-M. | | Deposit date: | 2010-05-17 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Allosteric non-bisphosphonate FPPS inhibitors identified by fragment-based discovery.
Nat.Chem.Biol., 6, 2010
|
|
3N52
 
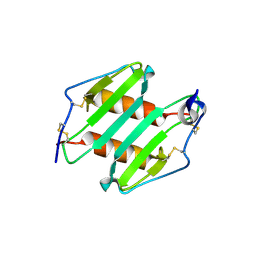 | | crystal Structure analysis of MIP2 | | Descriptor: | C-X-C motif chemokine 2 | | Authors: | Rajasekaran, D. | | Deposit date: | 2010-05-24 | | Release date: | 2011-06-08 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Model of GAG/MIP-2/CXCR2 Interfaces and Its Functional Effects.
Biochemistry, 51, 2012
|
|
3N5C
 
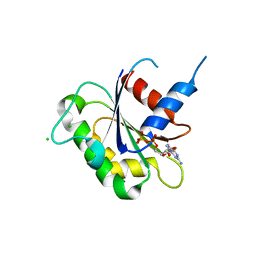 | | Crystal Structure of Arf6DELTA13 complexed with GDP | | Descriptor: | ADP-ribosylation factor 6, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Aizel, K, Biou, V, Cherfils, J. | | Deposit date: | 2010-05-25 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | SAXS and X-ray crystallography suggest an unfolding model for the GDP/GTP conformational switch of the small GTPase Arf6.
J.Mol.Biol., 402, 2010
|
|
3NBY
 
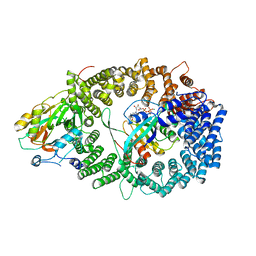 | | Crystal structure of the PKI NES-CRM1-RanGTP nuclear export complex | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4H5R
 
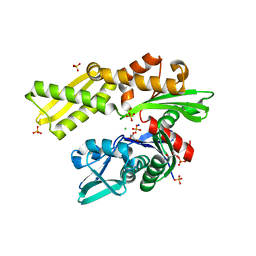 | | HSC70 NBD with Na, Cl and glycerol | | Descriptor: | CHLORIDE ION, GLYCEROL, Heat shock cognate 71 kDa protein, ... | | Authors: | Stec, B. | | Deposit date: | 2012-09-18 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | New crystal structures of HSC-70 ATP binding domain confirm the role of individual binding pockets and suggest a new method of inhibition.
Biochimie, 108, 2015
|
|
1HD7
 
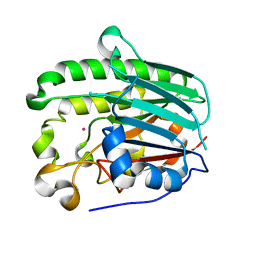 | | A Second Divalent Metal Ion in the Active Site of a New Crystal Form of Human Apurinic/Apyridinimic Endonuclease, Ape1, and its Implications for the Catalytic Mechanism | | Descriptor: | DNA-(APURINIC OR APYRIMIDINIC SITE) LYASE, LEAD (II) ION | | Authors: | Beernink, P.T, Segelke, B.W, Rupp, B. | | Deposit date: | 2000-11-09 | | Release date: | 2001-02-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Two Divalent Metal Ions in the Active Site of a New Crystal Form of Human Apurinic/Apyrimidinic Endonuclease, Ape1: Implications for the Catalytic Mechanism
J.Mol.Biol., 307, 2001
|
|
4HVU
 
 | |
3N1V
 
 | | Human FPPS COMPLEX WITH FBS_01 | | Descriptor: | (5-chloro-3-methyl-1-benzothiophen-2-yl)acetic acid, FARNESYL PYROPHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Rondeau, J.-M. | | Deposit date: | 2010-05-17 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Allosteric non-bisphosphonate FPPS inhibitors identified by fragment-based discovery.
Nat.Chem.Biol., 6, 2010
|
|
3N49
 
 | | Human FPPS COMPLEX WITH NOV_292 | | Descriptor: | FARNESYL PYROPHOSPHATE SYNTHASE, PHOSPHATE ION, naphtho[2,1-b]thiophen-1-ylacetic acid | | Authors: | Rondeau, J.-M. | | Deposit date: | 2010-05-21 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric non-bisphosphonate FPPS inhibitors identified by fragment-based discovery.
Nat.Chem.Biol., 6, 2010
|
|
3O9X
 
 | | Structure of the E. coli antitoxin MqsA (YgiT/b3021) in complex with its gene promoter | | Descriptor: | DNA (26-MER), GLYCEROL, Uncharacterized HTH-type transcriptional regulator ygiT, ... | | Authors: | Brown, B.L, Peti, W, Page, R. | | Deposit date: | 2010-08-04 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.0999 Å) | | Cite: | Structure of the Escherichia coli Antitoxin MqsA (YgiT/b3021) Bound to Its Gene Promoter Reveals Extensive Domain Rearrangements and the Specificity of Transcriptional Regulation.
J.Biol.Chem., 286, 2011
|
|
3OFW
 
 | | Crystal structure of recombinant Kunitz Type serine protease Inhibitor-1 from the Carribean sea anemone stichodactyla helianthus | | Descriptor: | CHLORIDE ION, Kunitz-type proteinase inhibitor SHPI-1 | | Authors: | Garcia-Fernandez, R, Redecke, L, Pons, T, Perbandt, M, Talavera, A, Gil, D, Gonzalez, Y, de los Angeles Chavez, M, Betzel, C. | | Deposit date: | 2010-08-16 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the recombinant BPTI/Kunitz-type inhibitor rShPI-1A from the marine invertebrate Stichodactyla helianthus.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3OGN
 
 | |
3M5D
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase K302R mutant complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reversible post-translational carboxylation modulates the enzymatic activity of N-acetyl-L-ornithine transcarbamylase.
Biochemistry, 49, 2010
|
|
